Abstract
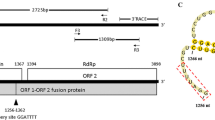
The complete genomic sequence of a variant of the recently reported maize-associated totivirus (MATV) from China was obtained from commercial maize in Ecuador. The genome of MATV-Ec (Ecuador) (4,998 bp) is considerably longer than that of MATV-Ch (China) (3,956 bp), the main difference due to a ≈ 1-kb-long capsid-protein-encoding fragment that is completely absent from the Chinese genome. Sequence alignments between MATV-Ec and MATV-Ch showed an overall identity of 82% at the nucleotide level, whereas at the amino acid level, the viruses exhibited 95% and 94% identity for the putative capsid protein and the RNA-dependent RNA polymerase (RdRp), respectively. Phylogenetic analysis of the viral RdRp domain indicated that MATV-Ec and MATV-Ch share a common ancestor with other plant-associated totiviruses, with Panax notoginseng virus A as the closest relative. MATV-Ec was detected in 46% (n = 80) of maize plants tested in this study, but not in endophytic fungi isolated from plants positive for the virus.


Similar content being viewed by others
References
Wickner RB, Ghabrial SA, Nibert ML, Patterson JL, Wang CC (2012) Totiviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz E (eds) Virus Taxon. Classif. Nomencl. viruses Ninth Rep. Int. Comm. Taxon. Viruses. Elsevier, San Diego, pp 639–650
Lachnit T, Thomas T, Steinberg P (2016) Expanding our understanding of the seaweed holobiont: RNA Viruses of the Red Alga Delisea pulchra. Front Microbiol 6:1–12. doi:10.3389/fmicb.2015.01489
Kondo H, Hisano S, Chiba S et al (2016) Sequence and phylogenetic analyses of novel totivirus-like double-stranded RNAs from field-collected powdery mildew fungi. Virus Res 213:353–364. doi:10.1016/j.virusres.2015.11.015
Martin RR, Tzanetakis IE, Sweeney M, Wegener L (2006) A virus associated with blueberry fruit drop disease. Acta Hortic. International Society for Horticultural Science (ISHS), Leuven, Belgium, pp 497–502
Guo L, Yang X, Wu W et al (2016) Identification and molecular characterization of Panax notoginseng virus A, which may represent an undescribed novel species of the genus Totivirus, family Totiviridae. Arch Virol 161:731–734. doi:10.1007/s00705-015-2716-4
Chen S, Cao L, Huang Q et al (2016) The complete genome sequence of a novel maize-associated totivirus. Arch Virol 161:487–490. doi:10.1007/s00705-015-2657-y
Abreu EFM, Daltro CB, Nogueira EOPL et al (2015) Sequence and genome organization of papaya meleira virus infecting papaya in Brazil. Arch Virol 160:3143–3147. doi:10.1007/s00705-015-2605-x
Liu H, Fu Y, Xie J et al (2012) Discovery of novel dsRNA viral sequences by in silico cloning and implications for viral diversity, host range and evolution. PLoS One 7:e42147. doi:10.1371/journal.pone.0042147
Morris TJ (1979) Isolation and analysis of double-stranded rna from virus-infected plant and fungal tissue. Phytopathology 69:854. doi:10.1094/Phyto-69-854
Froussard P (1992) A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res 20:2900
Quito-Avila DF, Ibarra MA, Alvarez R et al (2014) A raspberry bushy dwarf virus isolate from Ecuadorean Rubus glaucus contains an additional RNA that is a rearrangement of RNA-2. Arch Virol 159:2519–2521. doi:10.1007/s00705-014-2069-4
Huang C, Lu CL, Chiu H (2005) Structural bioinformatics A heuristic approach for detecting RNA H-type pseudoknots. 21:3501–3508. doi:10.1093/bioinformatics/bti568
Korabecna M (2007) The variability in the fungal ribosomal DNA (ITS1, ITS2, and 5.8 S rRNA gene): its biological meaning and application in medical mycology. Commun Curr Res Educ Top Trends Appl Microbiol 108:783–787. doi:10.1128/JCM.39.10.3617
Halgren A, Tzanetakis IE, Martin RR (2007) Identification, Characterization, and Detection of Black raspberry necrosis virus. Phytopathology 97:44–50. doi:10.1094/PHYTO-97-0044
Quito-Avila DF, Alvarez RA, Mendoza AA (2016) Occurrence of maize lethal necrosis in Ecuador : a disease without boundaries ? Eur J Plant Pathol. doi:10.1007/s10658-016-0943-5
Ghabrial SA, Suzuki N (2009) Viruses of plant pathogenic fungi. Annu Rev Phytopathol 47:353–384. doi:10.1146/annurev-phyto-080508-081932
White TJ, Bruns TD, Lee STJ (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Gelfand DH, Sninsky JJWT (eds) Innis MA. PCR Protoc. a Guid. to methods Appl. Academic Press, San Diego, California, pp 315–322
Sá Antunes TF, Amaral RJV, Ventura JA et al (2016) The dsRNA virus papaya meleira virus and an ssRNA virus are associated with papaya sticky disease. PLoS One 11:e0155240. doi:10.1371/journal.pone.0155240
Quito-Avila DF, Alvarez RA, Ibarra MA, Martin RR (2015) Detection and partial genome sequence of a new umbra-like virus of papaya discovered in Ecuador. Eur J Plant Pathol 143:199–204. doi:10.1007/s10658-015-0675-y
Acknowledgements
The authors would like to express their gratitude to maize growers for granting access to their fields and to researchers at INIAP-Portoviejo, especially to Alma Mendoza who helped with sample collection.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was partially funded by the Ecuadorean Science and Technology Secretariat (SENESCYT), through the National Emblematic Program PROMETEO.
Conflict of interest
All the authors declare they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Alvarez-Quinto, R.A., Espinoza-Lozano, R.F., Mora-Pinargote, C.A. et al. Complete genome sequence of a variant of maize-associated totivirus from Ecuador. Arch Virol 162, 1083–1087 (2017). https://doi.org/10.1007/s00705-016-3159-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-3159-2