Nanomaterials to combat SARS-CoV-2: Strategies to prevent, diagnose and treat COVID-19
- 1Laboratorio de Inmunología Celular y Viral, Unidad de Farmacología, Sección de Medicina, Facultad de Ciencias de la Salud, Universidad de La Laguna, San Cristóbal de La Laguna, Spain
- 2Research Unit, Hospital Universitario N. S. de Candelaria, Santa Cruz de Tenerife, Spain
- 3Genomics Division, Instituto Tecnológico y de Energías Renovables, Santa Cruz de Tenerife, Spain
- 4Servicio de Microbiología, Hospital Universitario N. S. de Candelaria, Santa Cruz de Tenerife, Spain
- 5Departamento de Análisis Matemático, Facultad de Ciencias, Universidad de La Laguna, Santa Cruz de Tenerife, Spain
- 6CIBER de Enfermedades Respiratorias, Instituto de Salud Carlos III, Madrid, Spain
- 7Faculty of Health Sciences, University of Fernando Pessoa Canarias, Las Palmas de Gran Canaria, Spain
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and the associated coronavirus disease 2019 (COVID-19), which severely affect the respiratory system and several organs and tissues, and may lead to death, have shown how science can respond when challenged by a global emergency, offering as a response a myriad of rapid technological developments. Development of vaccines at lightning speed is one of them. SARS-CoV-2 outbreaks have stressed healthcare systems, questioning patients care by using standard non-adapted therapies and diagnostic tools. In this scenario, nanotechnology has offered new tools, techniques and opportunities for prevention, for rapid, accurate and sensitive diagnosis and treatment of COVID-19. In this review, we focus on the nanotechnological applications and nano-based materials (i.e., personal protective equipment) to combat SARS-CoV-2 transmission, infection, organ damage and for the development of new tools for virosurveillance, diagnose and immune protection by mRNA and other nano-based vaccines. All the nano-based developed tools have allowed a historical, unprecedented, real time epidemiological surveillance and diagnosis of SARS-CoV-2 infection, at community and international levels. The nano-based technology has help to predict and detect how this Sarbecovirus is mutating and the severity of the associated COVID-19 disease, thereby assisting the administration and public health services to make decisions and measures for preparedness against the emerging variants of SARS-CoV-2 and severe or lethal COVID-19.
1 Introduction
In the last days of the year 2019, Chinese Health Authorities reported a cluster of twenty seven patients affected with “pneumonia of unknown etiology” (seven presenting sever symptoms), which appeared to be epidemiologically associated to a seafood and wet animal wholesale market in the city of Wuhan (Hubei Province, China) (Anderson et al., 2020; Bangaru et al., 2020; Cao et al., 2020). The microorganism responsible of these “pneumonia of unknown etiology” was rapidly identified in bronchoalveolar lavage samples collected from three patients of the Wuhan Jinyintan Hospital (30 December 2019) (Cao et al., 2020). In this regard, the first named 2019-nCoV virus (2019 novel coronavirus), later on named SARS-CoV-2 (severe acute respiratory syndrome coronavirus virus 2) (ICTV, 2020), was identified by local hospitals using a surveillance mechanism for “pneumonia of unknown etiology”. This mechanism was already established in the 2003 SARS outbreak crisis with the aim of allowing timely identification of novel pathogens and reaction in future outbreaks (Cao et al., 2020; International-Committee-on-Taxonomy-of-Viruses, 2020). Importantly, this surveillance system allowed the identification of some cases that were epidemiologically unrelated to Wuhan market outbreak (Ackermann et al., 2020; Ahmed et al., 2020; Bangaru et al., 2020; Cao et al., 2020; International-Committee-on-Taxonomy-of-Viruses, 2020).
The associated illness was first named “novel coronavirus-infected pneumonia” (NCIP) and the World Health Organization (WHO) recommended that the interim name of the agent causing the current outbreak should be 2019-nCoV and was considered the causative agent of the “pneumonia of unknown etiology” (Cao et al., 2020; Shimabukuro, 2021). This name complied with the WHO Best Practices for Naming of New Human Infectious Diseases, which were developed through a consultive process among partner agencies. On 30 January 2020, WHO declared that coronavirus disease 2019 (COVID-19) was a “public-health emergency of international concern (PHEIC)” (Amaechi et al., 2020). The name of the disease was provided by the WHO International Classification of Diseases (ICD; on 11 February 2020) as COVID-19, and the official name of the virus was given by the International Committee on Taxonomy of Viruses (ICTV; on 11 February 2020), as SARS-CoV-2 (ICTV, 2020). On 11 March 2000, the WHO declared the SARS-CoV-2/COVID-19 outbreak a global pandemic (Cucinotta and Vanelli, 2020; Shimabukuro, 2021).
The first complete sequences of the new CoV (SARS-CoV-2) genome obtained from Wuhan’s patients were submitted to GISAID (Global Initiative on Sharing All Influenza Data; https://www.gisaid.org/; (Liu et al., 2020d)) (Cao et al., 2020; Lu et al., 2020) and GenBank (accession number MN908947) where the virus strain was designated as Wuhan-Human 1 CoV (Wuhan-Hu-1 or WHCV) with a whole genome sequence of 29,903 nucleotides (nt) (Ackermann et al., 2020). At the same time, it was reported the epidemiological data of nine inpatients, from at least three hospitals in Wuhan, who were diagnosed with viral pneumonia of unidentified cause (Barton et al., 2020). Furthermore, some early cases were not epidemiologically associated with the market of Wuhan (Ahmed et al., 2020; Bangaru et al., 2020; International-Committee-on-Taxonomy-of-Viruses, 2020). The sequencing of bronchoalveolar lavage fluid samples of the above nine patients again identified the new CoV (Barton et al., 2020). More evidence for the presence of this CoV was obtained by virus isolation form the clinical specimens, viral culture, and cytotoxicity assays together with virus morphology analysis by electron microscopy (Barton et al., 2020; Cao et al., 2020), all of which are methods and devices that clearly use key nanotechnologies for SARS-CoV-2/COVID-19 detection and diagnosis (Caldas et al., 2020; Prasad et al., 2020; Akilesh et al., 2021; Jefferson et al., 2021; Sung et al., 2022).
The disease symptoms vary from person to person, from a paucisymptomatic or mild respiratory illness to an acute distress respiratory syndrome (ARDS). However, the prevalent reported symptoms associated with SARS-CoV-2/COVID-19 are fever, cough, fatigue, dyspnea (shortness of breath) and anosmia (partial or complete loss of the sense of smell), sore throat, headaches, ocular manifestations, chest pain and diarrhea. It also includes abnormal coagulation and lymphopenia that together with cytokine storm, hyper-inflammation, sepsis and septic shock accompany severe pneumonia, ARDS and multiorgan dysfunction, including neuropathogenesis, liver, kidney and heart failure (Bizzarro et al., 2011; Ackermann et al., 2020; Ahmed et al., 2020; Amaechi et al., 2020; Arslan et al., 2020; Baig et al., 2020; Bangaru et al., 2020; Barton et al., 2020; Brann et al., 2020; Cao et al., 2020; Chan et al., 2020; Chen et al., 2020; Cheung et al., 2020; China-National-Health-Commission-(Beijing), 2020; Corbett et al., 2020; Devaux et al., 2020; Diao et al., 2020; Gavriatopoulou et al., 2020; Gibson et al., 2020; Goyal et al., 2020; Guan et al., 2020; Hariri and Hardin, 2020; Helms et al., 2020; International-Committee-on-Taxonomy-of-Viruses, 2020; Kuster et al., 2020; Lechien et al., 2020; Ledford, 2020; Levi et al., 2020; Lipsitch et al., 2020; Mazzoni et al., 2020; Menter et al., 2020; Ouyang et al., 2020; Puelles et al., 2020; Ruan et al., 2020; Seah and Agrawal, 2020; Toscano et al., 2020; Tsivgoulis et al., 2020; Verdoni et al., 2020; Baden et al., 2021; Shimabukuro, 2021). Therefore, COVID-19 infections affect various organs in the body, including the central nervous system (Baig et al., 2020; Wu et al., 2020), cardiovascular system (Zheng et al., 2020), kidneys (Cheng et al., 2020b), gastrointestinal tract (Wong et al., 2020b), and liver (Zhang et al., 2020a; Bangash et al., 2020) (Figure 1A). Furthermore, a common cause of death is an uncontrolled hyperinflammatory response (i.e., with uncontrolled cytokine levels), which can lead to stroke due to blood clots, organ failure, and heart attacks (Hasanzadeh et al., 2021). Since the SARS-CoV-2 must bind to angiotensin-converting enzyme 2 (ACE2) to enter and infect host cells in humans (Zhou et al., 2020a; Shang et al., 2020b; Walls et al., 2020b; Wang et al., 2020e; Lan et al., 2020), the expression (partly determined by the stage of development) and body localization of ACE2 are important to understand COVID-19 symptoms and disease (Hamming et al., 2004; Yang et al., 2020b; Ni et al., 2020) (Figure 1A). Noteworthy, ACE2 is a zinc metallopeptidase involved in regulation of blood pressure and cardiac function (Tikellis and Thomas, 2012). It has recently been reported that SARS-CoV-2-targeted ACE2-expressing organs lead to severe pathobiological manifestations followed by multiple organ failure (Salamanna et al., 2020) (Figure 1A, scheme representing the different organs and tissues that express ACE2, and could be infected by SARS-CoV-2). The ACE2 receptor for SARS-CoV-2 infection was previously reported to be the receptor for the Sarbecovirus SARS-CoV-1 (Li et al., 2003) and for the human coronavirus (HCoV)-NL63 (Hofmann et al., 2005), an alphacoronavirus known to cause mild upper respiratory tract infection. The entry steps of the SARS-CoV-2 viral particles are mediated by the external S glycoprotein, which is arranged as a homotrimers (peplomers) (Cavanagh, 1995) that give the characteristic Sun’s corona-like aspect to the viral particles (Liu et al., 2020b; Liu et al., 2020c) (see images at NIH (NIAID) website “https://www.flickr.com/photos/niaid/albums/72157712914621487”), therefore classified as the coronaviridae family (de Groot et al., 1987). SARS-CoV-2 fusion with cell-plasma membrane and cell entry are therefore initiated by the interaction of the receptor binding domain (RBD) of the S protein to human ACE2 at the cell-surface (Li et al., 2003; Li et al., 2005; Yan et al., 2020a; Wang et al., 2020e; Wan et al., 2020) (Figure 1B, scheme representing key viral and cell molecules involved in virus attachment and fusion with target cell that mediate viral entry and infection). Then, the S protein, anchored to ACE2 in its prefusion state (Figure 1B), undergoes proteolytic cleavage catalyzed by several host proteases, such as furin that cleaves at the polybasic region (RRAR685|S), and TMPRSS2 (transmembrane serine protease 2) that recognizes the canonical S1/S2 cleavage site, as well as other sites at the S protein (Hoffmann et al., 2020a; Bestle et al., 2020; Hoffmann et al., 2020b; Wang et al., 2020e; Fraser et al., 2022) (Figure 1B). Likewise, the S protein could be also cleaved by cathepsin B/L, within endosomes after virus ACE2-mediated endocytosis, promoting virus-endosome membrane fusion (Hoffmann et al., 2020b; Bollavaram et al., 2021; Ou et al., 2021) (Figure 1B, Inbox scheme). All these proteolytic cleavages expose the fusion peptide of the S2 subunit of the virus, facilitating virus-cell membranes fusion (Figure 1B) and the entry of viral RNA + genome into the cell, starting the SARS-CoV-2 infection process (reviewed in (Peng et al., 2021a; Zhang et al., 2021a; Jackson et al., 2022)). Furin could also cleave the S protein into the S1 and S2 subunits, which remain associated, during the late steps of the viral cycle in virus-producing cells (Hoffmann et al., 2020a; Shang et al., 2020a). This event leads to new viral particles that easily expose the S2-fusion peptide favoring cell-cell spread and cell infection (Hoffmann et al., 2020a; Shang et al., 2020a; Papa et al., 2021) (Figure 1B, Bottom scheme). The furin cleavage site at the S protein is thought to be responsible for the virulence of SARS-CoV-2 (Hoffmann et al., 2020a; Johnson et al., 2020; Johnson et al., 2021). Furthermore, several cofactors have been involved in SARS-CoV-2 entry and infection (Figure 1B, Top scheme). Thus, Neuropilin-1 (NRP1) appears to trigger SARS-CoV-2 viral entry and infection (Cantuti-Castelvetri et al., 2020; Daly et al., 2020). NRP1 has been proposed to mediate SARS-CoV-2 entry into the brain via the olfactory bulb (Cantuti-Castelvetri et al., 2020), since NRP1 is expressed in respiratory and olfactory epithelia (Cantuti-Castelvetri et al., 2020; Daly et al., 2020). Moreover, high-density lipoprotein (HDL) scavenger receptor B type 1 (SR-B1) has been reported to promote SARS-CoV-2 entry, in an ACE2-dependent manner (Wei et al., 2020), whereas the transmembrane glycoprotein CD147 (also named BSG (basigin (Ok blood group) (Kaname et al., 1993) or EMMPRIN (extracellular matrix metalloproteinase inducer) (Biswas et al., 1995)), could mediate viral entry and infection by endocytosis, even in cells lacking ACE2 expression (Wang et al., 2020c). Moreover, it seems that CD147 and ACE2 cooperate during SARS-CoV-2 infection, since ACE2 levels are modulated by CD147 density and they are coregulated by viral infection (Fenizia et al., 2021).
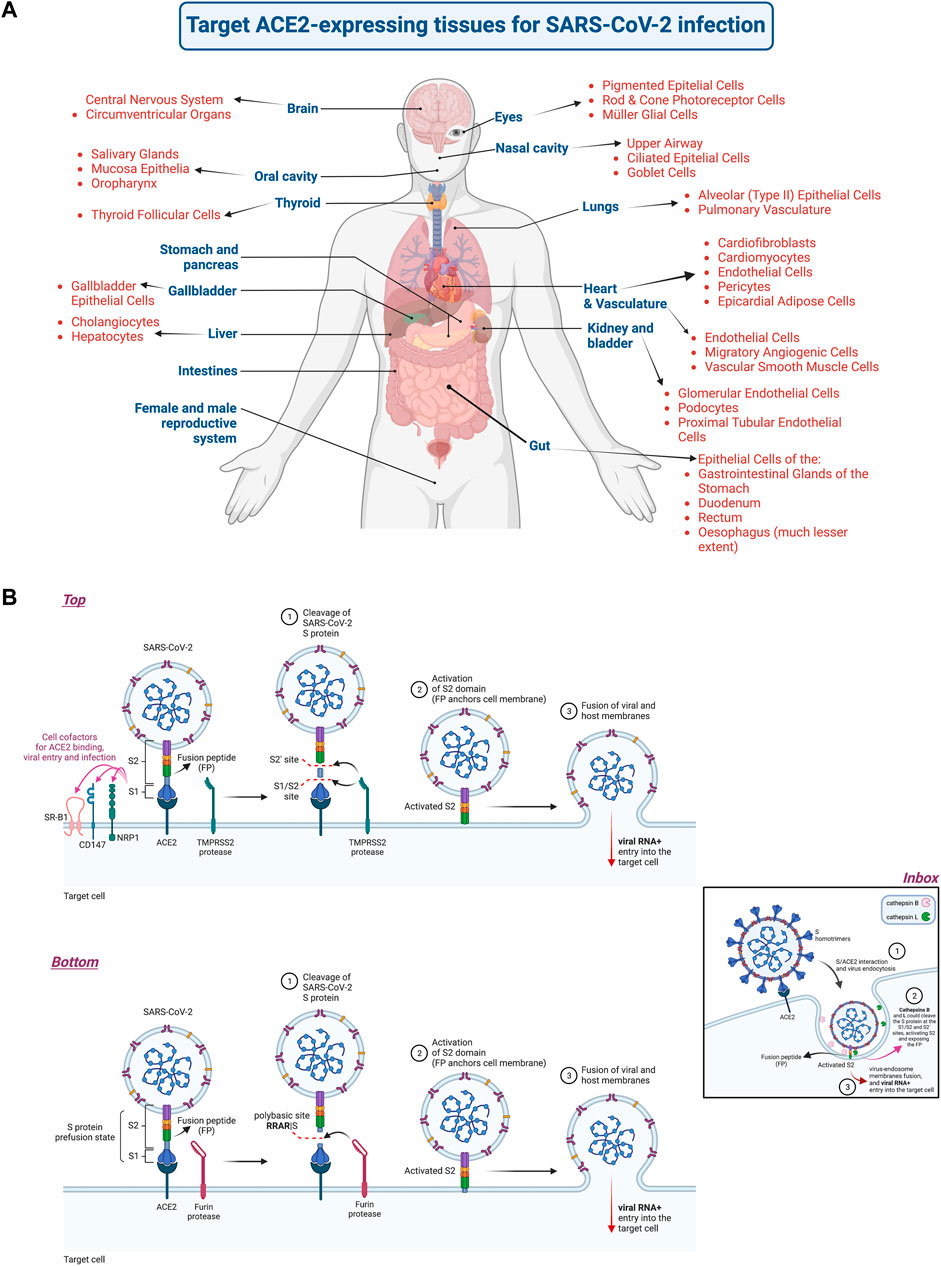
FIGURE 1. Cells, tissues and organs that express the ACE2 receptor and are infected by SARS-CoV-2, and the mechanisms of early SARS-CoV-2 infection. (A) Scheme representing cells, tissues and organs expressing the ACE2 receptor that allows the SARS-CoV-2 virus to infect them, thereby being associated with tissue damage and organ failure. (B) Scheme representing the main viral and cellular molecules involved in SARS-CoV-2 attachment and fusion with target cell that mediate viral entry and infection. The S protein (with its S1 and S2 subunits in the prefusion state) interacts with ACE2 leading to conformational changes in the protein that facilitate the proteolytic cleavage of the S protein that further exposes the fusion peptide (FP). This S cleavage is catalyzed by several host proteases, such as TMPRSS2 that recognizes the canonical S1/S2 cleavage site, as well as other sites at the S protein (e.g., S2’) (Top scheme), and furin that excises at the polybasic region (RRAR|S) located between S1 and S2 subunits, and near the S2-fusion peptide (Bottom scheme). The S protein could also be cleaved by the endosome enzymes cathepsin B/L, after ACE2-mediated endocytosis of the virus, promoting virus-endosome membranes fusion (Inbox scheme). All these proteolytic cleavages expose the fusion peptide (FP) of the S2 subunit of the virus, facilitating virus-cell membranes fusion and the entry of viral RNA + genome into the cell, starting the SARS-CoV-2 infection process. Several cofactors have been reported to be involved in SARS-CoV-2 entry and infection, such as NRP1, reported to mediate SARS-CoV-2 entry into the brain via the olfactory bulb, the SR-B1 receptor, proposed to promote SARS-CoV-2 entry (in an ACE2-dependent manner) and the transmembrane glycoprotein CD147 (BSG or EMMPRIN) that could mediate viral entry and infection by endocytosis cooperating with ACE2 (Top scheme). Designs and templates are created with BioRender.
Therefore, nano-based strategies to battle SARS-CoV-2/COVID-19 must confront all symptoms of the disease, particularly to avoid severe outcomes, and to neutralize the infection of cells from different tissues and organs that could be targeted by the virus, through vaccine-elicited specific and optimal immune responses, mainly driven against the S viral immunogen.
In this scenario, viral isolation, culture and sequencing were key methods, based on nanotechnology and nano-reagents or materials that help to achieve fasts diagnosis and to endorse and develop urgent public health measures and responses, as the WHO preparedness for emerging epidemic threats recommends (McCloskey et al., 2014; Elbe and Buckland-Merrett, 2017; Jain et al., 2018; Kluge et al., 2018; Global-Preparedness-Monitoring-Board-(GPMB), 2019; Baden et al., 2021). These nano-based tools and strategies allowed the detection of a new RNA+ genome from patients’ samples, and the identification of the new SARS-CoV-2 virus of the lineage B of the genus betacoronavirus (Cao et al., 2020). In general, nanomaterials and nanotechnological approaches to combat viral infections consist in diagnostic sensors, vaccine formulations, and protective coatings, and vehicles for delivery of anti-viral vaccines and adjuvants, as well as for new treatments to palliate COVID-19 associated symptoms and to limit disease severity (reviewed in (Campos et al., 2020; Cavalcanti and Cajubá De Britto Lira Nogueira, 2020; Chung et al., 2020; Gupta et al., 2020; Pisani et al., 2020; Talebian and Conde, 2020; Weiss et al., 2020; Asdaq et al., 2021; Constantin et al., 2021; Petkar et al., 2021; Rashidzadeh et al., 2021; Varahachalam et al., 2021; Goharshadi et al., 2022; Souri et al., 2022)). Nanoreagents and derived materials have intrinsic antipathogenic properties that are able to inactivate and degrade the SARS-CoV-2 virus, as well as other viruses, bacteria and microorganisms via generation of reactive oxygen species in contact with the virus or triggered by photocatalysis. We have considered materials used in face masks and protective personal equipment (PPE) for prevention. These nanomaterials could offer new tools and disinfection methods in healthcare facilities. Vehicles used for drug delivery are similar to those used for vaccine-associated immunogen delivery, which could function as adjuvants for immune system activation (reviewed in (Petkar et al., 2021)). We have considered nano-based vaccines as the real treatment for prevention and treatment of SARS-CoV-2 infection and COVID-19 disease. We have summarized the main diagnoses tests used (serological and RT-qPCR, useful in point-of-care facilities) (Valera et al., 2021) and emphasizing genome-mediated approximations based on nanopore technology (Abraham et al., 2008; Ciuffreda et al., 2021b; Liefting et al., 2021; Lin et al., 2021; Ju et al., 2022), an outstanding electrochemical biosensor on nanoscale used for SARS-CoV-2 detection and sequencing (Yakovleva et al., 2022), and viral whole genome sequencing (WGS) (Gohl et al., 2020; Rosenthal et al., 2022). These strategies are considered high performance nanotechniques for virus genome sequencing, diagnosis and virus surveillance, where their utility to combat SARS-CoV-2/COVID-19 have been clearly demonstrated in this pandemic crisis. All these nano-based designs, tools and strategies have allowed a historical, unprecedent, real time epidemiological surveillance, diagnosis of SARS-CoV-2 infection and transmission, at community and international levels, predicting how this Sarbecovirus is mutating and the severity of the associated COVID-19 disease.
In this review we aimed to address all these nano-based strategies to battle SARS-CoV-2/COVID-19 that assist the administration and public health services to make decisions and measures for preparedness against the emerging variants of SARS-CoV-2 and severe or lethal COVID-19.
2 Nanomaterials in the battle against SARS-CoV-2: Physical barriers, vaccines, genomics-based virosurveillance and diagnosis
2.1 Physical barriers for prevention: Face masks and protective personal equipment
The entire world has witnessed that prevention strategies against the still ongoing COVID-19 pandemics have included vaccines, antiviral drugs and, since the beginning, personal protective countermeasures, such as facemasks together with physical distancing (Chu et al., 2020; Brainard et al., 2020; Li et al., 2021b; Raman et al., 2021; Rowan and Moral, 2021). In fact, nanomaterials and associated technologies are also crucial to control person-to-person virus transmission and viral community spreading (i.e., face masks, sanitizers and disinfectants, drugs and vaccines) (Barycka et al., 2020; Chu et al., 2020; Brainard et al., 2020; Carraturo et al., 2020; Ilyas et al., 2020; Asdaq et al., 2021; Li et al., 2021b; Dhama et al., 2021; Hodgson et al., 2021; Meo et al., 2021; Rohilla, 2021; Rowan and Moral, 2021; Salian et al., 2021; Tabatabaeizadeh, 2021; Thompson et al., 2021; Vitiello et al., 2021; Kianpour et al., 2022). If these measures to control transmission are neglected it will lead to the emergence of a plethora of new variants of the SARS-CoV-2 virus [including the variants of concerns (VOCs)] with may associate with increased infectious and transmission properties that compromise natural and vaccine-elicited immune responses and protection, as well as the effectiveness of the assays for rapid detection and diagnosis (Alcoba-Florez et al., 2020a; Weisblum et al., 2020; Bal et al., 2021; Bian et al., 2021; Boehm et al., 2021; Camp et al., 2021; Liu et al., 2021d; Hoffmann et al., 2021; Raman et al., 2021; Salleh et al., 2021; Singh et al., 2021; Tao et al., 2021; Alejo-Cancho et al., 2022; Mertens et al., 2022; Sonnleitner et al., 2022).
The main transmission route of SARS-CoV-2 from person to person is aerosol (Morawska and Cao, 2020; Setti et al., 2020), among respiratory droplets, oral-fecal transmission and contact (i.e., fomites), bloodborne, mother-to-child (reported to occur at low rates) and animal-to-human transmission (Ackermann et al., 2020; Ahmed et al., 2020; Amaechi et al., 2020; Arslan et al., 2020; Aydillo et al., 2020; Bangaru et al., 2020; Bonato et al., 2020; Brann et al., 2020; Cao et al., 2020; Carvalho et al., 2020; Chang et al., 2020; Cheung et al., 2020; Dona et al., 2020; Eslami and Jalili, 2020; Grassia et al., 2020; Guan et al., 2020; Guo et al., 2020c; Hoffmann et al., 2020b; Huang et al., 2020a; International-Committee-on-Taxonomy-of-Viruses, 2020; Jin et al., 2020; Khanh et al., 2020; Lechien et al., 2020; Lednicky et al., 2020; Lescure et al., 2020; MacIntyre and Ananda-Rajah, 2020; Mondelli et al., 2020; Nishiyama et al., 2020; Nzediegwu and Chang, 2020; Pastorino et al., 2020; Poggio et al., 2020; Rothe et al., 2020; Santarpia et al., 2020; Shereen et al., 2020; van Doremalen et al., 2020; Voskarides, 2020; Wang et al., 2020f; Xu et al., 2020a; Al-Sharif et al., 2021; Baden et al., 2021; Birkhead et al., 2021; Chan et al., 2021; Chaubey et al., 2021; Chaudhary et al., 2021; Conti et al., 2021; Cuicchi et al., 2021; Dumitriu et al., 2021; Kumar et al., 2021; Kutter et al., 2021; Musa et al., 2021; Peacock et al., 2021a; Sawatzki et al., 2021; Shimabukuro, 2021; Valencak et al., 2021; Yang et al., 2021a; Zaneti et al., 2021; Chan et al., 2022; Costa et al., 2022; de Oliveira-Filho et al., 2022; Jairak et al., 2022; Kang et al., 2022; Katona et al., 2022; Kyle et al., 2022; Patane et al., 2022; Pecora et al., 2022; Wake, 2022; WHO, 2022). The average size of this virus is 60–140 nm with a volume of about 106 nm3 (Bar-On et al., 2020; Zhu et al., 2020c). The likelihood of airborne transmission is very low unless droplets (less than 5 μm in diameter) are contaminated with the virus and remain in the air for long periods. The air transports exhaled droplets that might contain the virus, when expired, the liquid content evaporates, diminishing their size. These small droplets are free to travel in the air, thereby being easily transported by air currents, carrying their viral content several meters from where they originated and occupying the entire volume of a room (Bourouiba, 2020; Setti et al., 2020). SARS-CoV-2 is highly stable in aerosol and on surfaces compared to SARS-CoV-1, maintaining the virus infectious for hours in the aerosol (Anfinrud et al., 2020; Morawska and Cao, 2020; Paules et al., 2020; van Doremalen et al., 2020; Ong et al., 2021). These facts imply that in the absence of face masks the distance of 1–2 m among people is not enough to safeguard from SARS-CoV-2 infection risk (Parshina-Kottas et al., 2020), as it has been reported to happen in hospitals (i.e., SARS-CoV-2 RNA has been detected in air samples collected inside the hospitals, thereby the airborne route has to be considered an important pathway for contamination), schools and other indoors spaces (Liu et al., 2020f; Nissen et al., 2020; Santarpia et al., 2020; Akhmetzhanov et al., 2021; Chau et al., 2021; Ding et al., 2021; Kumar et al., 2021; Kumari et al., 2021; Leeman et al., 2022; Marchese et al., 2021; Miyoshi et al., 2021; Murewanhema et al., 2021; Ong et al., 2021; Salmenjoki et al., 2021; Sami et al., 2021; San et al., 2021; Jung et al., 2022; Kirsten et al., 2022; Ladhani et al., 2022; Leng et al., 2022; Nagy et al., 2022; Polechova et al., 2022; Viner et al., 2022; White et al., 2022). Therefore, SARS-CoV-2 is transmitted by bioaerosols (<10 μm) and droplets (>10 μm) projected during breathing, speaking and coughing (Klompas et al., 2020a; Binder et al., 2020; Liu et al., 2020f; Nissen et al., 2020; van Doremalen et al., 2020; Zhang and Duchaine, 2020; Alsved et al., 2022), with an estimation of about 4.8×105 virus gene copies/mL in bioaerosol samples (Cevik et al., 2021; Malik et al., 2021; Johnson et al., 2022).
On the other hand, we must consider that with SARS-CoV-2, it has been shown that contagiousness commonly occurs before developing any symptom (Arons et al., 2020; Zhou et al., 2020b; Le et al., 2020; Mirjalali et al., 2020; Moghadas et al., 2020; Bae et al., 2021; Muller, 2021; Murata et al., 2021; Wilmes et al., 2021; Fakhruddin et al., 2022; Mugglestone et al., 2022; Overton et al., 2022; Pettifor et al., 2022; Rajme-Lopez et al., 2022), compared with seasonal flu, for example, where most transmissions occur after a person has developed symptoms (Leung et al., 2015b; Ip et al., 2017; Furuya-Kanamori and Yakob, 2018). The average SARS-CoV-2 incubation period is thought to be about 5 days, while peak infectiousness begins 2 days before symptoms reveal themselves (Yan et al., 2020b; Zhou et al., 2020b; Hua et al., 2020; McAloon et al., 2020; Noh et al., 2020; Tan et al., 2020; Bae et al., 2021; Murata et al., 2021; Yan et al., 2021). Moreover, viral loads in asymptomatic and symptomatic infected individuals have been reported to be similar (Arons et al., 2020; Lee et al., 2020b; Zhou et al., 2020b; Hoehl et al., 2020; Kimball et al., 2020; Kociolek et al., 2020; Le et al., 2020; Sohn et al., 2020; Uhm et al., 2020; Zou et al., 2020; Hasanoglu et al., 2021; Ra et al., 2021; Sah et al., 2021; Zuin et al., 2021), as the efficiency to transmit the virus (Li et al., 2020a; Arons et al., 2020; Wong et al., 2020a; Bae et al., 2020; Gao et al., 2020b; Kim et al., 2020b; Li et al., 2020b; Qian et al., 2020b; Zhou et al., 2020b; Day, 2020; Wang et al., 2020g; Hao et al., 2020; He et al., 2020; Jiang et al., 2020; Moghadas et al., 2020; Rivett et al., 2020; Sohn et al., 2020; Glenet et al., 2021; Hasanoglu et al., 2021; Joshi et al., 2021; Passarelli et al., 2021; Sah et al., 2021). Hence, the majority of infections occur pre-symptomatically, with the infected people ignoring that they have the disease (Ferretti et al., 2020; He et al., 2020; Moghadas et al., 2020).
In this pandemic scenario, the most basic method of prevention against COVID-19 is therefore to wear face masks (Howard et al., 2021; Ju et al., 2021). The face mask must be worn by either non-infected or infected people (Figure 2A). Thus, in infected individuals, facemasks could prevent the aerosol spread of the virus to other people, whereas the mask could protect against SARS-CoV-2 in non-infected individuals by covering their faces (Pezzini and Padovani, 2020; Catching et al., 2021). Face masks retain on their surface of the textile the virus and the constant use of this PPE (protective personal equipment) forces to renew the mask constantly, particularly in people who are exposed to the virus as in shops, pharmacies, schools, administration buildings, offices and hospitals, among other professions and sensitive infrastructures to reduce viral transmission and infection (Beesoon et al., 2020; Sanchez et al., 2020; Forum, 2020; Gholami et al., 2021; Howard et al., 2021; Ingram et al., 2021; Lessler et al., 2021). N95 and filtering facepiece 2 and 3 (FFP2 and FFP3) respirators that can filter from 95 to 99% of airborne particles, respectively, and surgical masks (filters around 80% of contaminants of certain size) are the most common face masks used during COVID-19 pandemics which provide protection against the airborne virus and larger infectious particles (Bartoszko et al., 2020; Karim et al., 2020; Sommerstein et al., 2020; Clinkard et al., 2021; Dheda et al., 2021; Regli et al., 2021; Rohit et al., 2021) (Figure 2A). These face masks have also been improved by using nanomaterials that present antimicrobial and antiviral properties, such as nanoparticles (NPs) (i.e., melt-blown polypropylene and nylon-cotton with antiviral zinc oxide-NPs) (Gonzalez et al., 2021), metal NPs (i.e., silver NPs (AgNPs)) (Valdez-Salas et al., 2021), spray or polymers of copper, gold, silver, zinc oxide (this could be washed and reused) or TiO2 (TiO2Ag-based facemasks are able to degrade 99.99% of viruses under zero light conditions) (Balagna et al., 2020; Konda et al., 2020; Mahapatra et al., 2020; Nickels, 2020; Sportelli et al., 2020; Wang et al., 2021a; Nbic+, 2022; Promethean-Particles-Ltd, 2022; Sonovia-SonoMask, 2022), in order to stop the virus transmission chain (Figure 2A). These metal oxide-derived NPs, embedded in face masks, act as antiviral or, in general, as antimicrobial compounds by inhibiting the catalytic activity of the microorganisms’ enzymes (i.e, viral polymerases and proteinases), which are key for their survival and pathogenicity (Cai et al., 2019; Wang et al., 2021a; Melk et al., 2021; Pachaiappan et al., 2021; Mendes et al., 2022). Moreover, due to their small size, these metal oxide-derived NPs enhance the surface contact with viruses and bacteria, as reported for Ag-TiO2 single atom nanozyme (SAN) that allows efficient adsorption for SARS-CoV2 through the association of Ag atoms with Cys and Asn residues of the RBD sequence of the viral S1 subunit (Wang et al., 2021a). These metal oxide-derived NPs, in contact with virus could also trigger the formation of reactive oxygen species (i.e., hydrogen peroxide anions and superoxide radical anions) which cause damage to the viral membranes and cell wall of bacteria, as well as to internal components, thereby impairing their uncoating, growth and integrity (Cai et al., 2019; Hamza et al., 2021; Melk et al., 2021; Pachaiappan et al., 2021; Mendes et al., 2022). Noteworthy, the effects of these metal oxide-derived NPs could depend on their composition and dose (Zanet et al., 2019; Wang et al., 2021a). Therefore, the use of these NPs, polymers and sprays for face masks production or treatment implies accurate techniques and procedures, in order to assure the maximal protection of face masks against SARS-CoV-2.
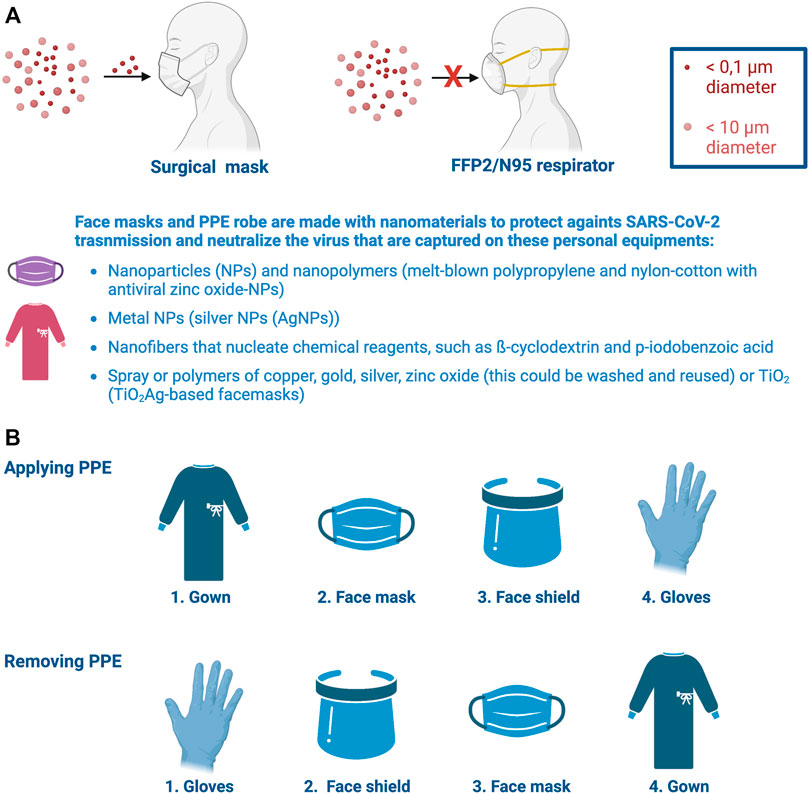
FIGURE 2. Nanomaterials used for protective personal equipment. (A) FFP2/NP95 masks are the appropriate protective respirators against this virus mainly transmitted by aerosol compared with surgical masks. Face masks and PPE robes have been incorporated nanoreagents and nanomaterials to improve personal protection against SARS-CoV-2 airborne infection and transmission to neutralize the virus in contact with these personal equipments. These nanomaterials are indicated in panel (A). (B) Personal training and correct protocols for wearing (applying) and removing these PPEs from the body are critical to ensure right protection of health personnel against SARS-CoV-2 infection. Designs and templates are created with BioRender.
All these nanoparticles are also used to produce PPE robe (Reddy et al., 2019; Bauchner et al., 2020; JH et al., 2020; Ortega et al., 2020) (Figure 2B), where PPE innovation will also help to overcome the important COVID-19 PPE waste challenge (IFC, 2021). Face masks and PPE robes have been also improved to fight and prevent COVID-19/SARS-CoV-2 infection by using nanofibers to create a dense weblike surface area against infection in the PPEs (Figure 2). These webs of nanofibers could nucleate some chemical reagents, such as β-cyclodextrin and p-iodobenzoic acid that allow nanofibers to act against SARS-CoV-2, thereby degrading the virus and reducing the risk of inhaling the virus or to be contaminated by contact with masks or PPE robes carrying the virus (Ramaseshan et al., 2006; Kadam et al., 2021; Rasmi et al., 2021; Owida et al., 2022). The nanofibers could be also presented in a variety of layers, including a water repellent and a skin-friendly silk layer with a 98% filtration guide, leading to an efficient protective PPE mask or robe that avoid SARS-CoV-2 transmission and infection (Chaudhary et al., 2021). Moreover, it has been reported that SARS-CoV-2 can remain on fomites for more than 9 days and potentially infectious (Kampf et al., 2020b; Bueckert et al., 2020; Hosseini et al., 2021; Marcenac et al., 2021). Therefore, several nanomaterials have been used to disinfect or remove this Sarbecovirus from different contaminated surfaces, such natural NPs-, metal NPs- and nanopolymers-based disinfectants (reviewed in (Souri et al., 2022)).
2.2 COVID-19 vaccines by nano-based strategies: Prevention and treatment
Physical barriers are required and are therefore very useful to protect ourselves and the others from infection, contagion and community spreading of the virus, lowering the concomitant emergence of viral variants (Střížová et al., 2020; Swain, 2020; Mello et al., 2022), which threatens individual and community immunity that might have been generated during previous infections or by vaccines (Arora et al., 2021; Boehm et al., 2021; Caniels et al., 2021; Arora et al., 2022; Salehi-Vaziri et al., 2022), as it has happened with the appearance of several Omicron VOCs (Cao et al., 2022; Flemming, 2022; Hachmann et al., 2022). Therefore, vaccines are the tools to elicit immunity against SARS-CoV-2 infection, reinfection by VOCs and severe COVID-19 disease, where face masks enhance vaccine efficiency in this crucial community and worldwide task. The community use of face masks therefore lower both virus transmission rates and the severity of the disease allowing to save more lives (Klompas et al., 2020b; Esposito et al., 2020; Gandhi and Rutherford, 2020; Leung et al., 2020). Likewise, it should be considered that combining vaccination and face masks is much less costly to apply than the use of a plethora of non-efficient drugs together with hospital intensive cares measures which are associated with high economic, social and live costs (Beesoon et al., 2020; Esposito et al., 2020; George et al., 2020; Grasselli et al., 2020; Hill et al., 2020; Iannaccone et al., 2020; Lerner et al., 2020; Rosenthal et al., 2020; Shaffer, 2020; Singh et al., 2020; Cleary et al., 2021; Di Fusco et al., 2021; Kompaniyets et al., 2021; Ohsfeldt et al., 2021; Bartsch et al., 2022; Brüssow and Zuber, 2022; Popescu et al., 2022).
Before presenting the different vaccine designs used against SARS-CoV-2/COVID-19, we summarize the immune responses elicited against SARS-CoV-2 infection and how the virus pervert immune protection leading to the COVID-19 pathology.
The innate immune responses are rapidly developed against this Sarbecovirus which very early during the infection leads to the production of type I interferons (IFNs), key antiviral cytokines that induce a large range of proteins that impair viral replication in infected cells (Lokugamage et al., 2020; Madden and Diamond, 2022). SARS-CoV-2 evades the host innate immune system by reducing IFN levels, its mediated signals and activation pathways through different SARS-CoV-2 proteins (Shin et al., 2020a; Lei et al., 2020; Miorin et al., 2020; Xia et al., 2020; Wang et al., 2021c; Wang et al., 2021d; Hayn et al., 2021; Kimura et al., 2021; Min et al., 2021; Munnur et al., 2021; Rashid et al., 2021; Vazquez et al., 2021; Chiale et al., 2022; Oh and Shin, 2022; Osipiuk et al., 2022; Znaidia et al., 2022). Moreover, genetic and immunological defects in the type-I IFN responses are known to cause fatal outcomes in otherwise healthy SARS-CoV-2 infected patients (Zhang et al., 2020d; Zhang et al., 2022a). Likewise, SARS-CoV-2 escape the translation inhibition as a result of some viral proteins (i.e., Nsp14 with the concert of Nsp10) (Hsu et al., 2021) and mRNAs that elude translation inhibition by a stem loop structure in the 5ʹ untranslated region (UTR) of all viral transcripts (Tidu et al., 2020).
Adaptative immune responses against SARS-CoV-2 antigens are important to control viral infection and the severity of the COVID-19 disease (Rydyznski Moderbacher et al., 2020). Specific and effective CD8+ T cell response is associated with viral clearance and mild disease (Bergamaschi et al., 2021; Notarbartolo et al., 2021). In acute COVID-19, effective CD8+ and CD4+ T cell responses are associated with positive clinical outcomes (Diao et al., 2020; Su et al., 2020; Notarbartolo et al., 2021; Rha et al., 2021). This T cell activation is also observed in asymptomatic SARS-CoV-2 infected individuals (Grau-Exposito et al., 2021; Le Bert et al., 2021). However, exacerbated T cell responses could exhaust these immune cells, being associated with poor clinical outcomes (Lucas et al., 2020b; Diao et al., 2020; Mathew et al., 2020; Song et al., 2020; Su et al., 2020), particularly in patients presenting comorbidities (Lucas et al., 2020b; Yu et al., 2021). In fact, humoral response has been reported to be retarded in patients with severe COVID-19 and in fatal outcomes (Lucas et al., 2021). Follicular T and B cell cooperation in germinal centers (GCs) are key to create specific memory cells against viral antigens and humoral and cellular long-lasting repertoires and responses (Allen et al., 2007; Kräutler et al., 2017; Cyster and Allen, 2019). Severe SARS-CoV-2 infection and illness have been associated with the absence of GCs (Kaneko et al., 2020) as observed by the decrease in the number of T follicular cells. This could be in agreement with the robust extrafollicular B cell responses with concomitant increase in proinflammatory cytokine levels and the neutralizing antibody titers observed in severe COVID-19 patients with critical pneumonia (Woodruff et al., 2020), where delayed production and responses of specific neutralizing antibodies (nAbs) (Lucas et al., 2021) and the pre-existence of type-I IFN auto-Abs (Bastard et al., 2020; Bastard et al., 2021; Wang et al., 2021b) have been reported.
Therefore, effective vaccine designs against SARS-CoV-2 infection and COVID-19 disease should induce robust and persistent GC reactions, in order to generate high-affinity and durable neutralizing antibody responses together with T cell memory coordinated responses and reservoir (Brouwer et al., 2020; Jagannathan and Wang, 2021; Teijaro and Farber, 2021; Turner et al., 2021; Winkler et al., 2021). Innovative nanovaccine formulations, as we further describe, have demonstrated to successfully achieve optimal activation of both innate and adaptive immune responses against SARS-CoV-2 infection (i.e., mRNA vaccines-mediated GC-B cell response to SARS-CoV-2 (Lederer et al., 2020; Turner et al., 2021; Laidlaw and Ellebedy, 2022)), in order to achieve herd immunity (reviewed in (Shin et al., 2020b; Chauhan et al., 2020; Chung et al., 2020; Constantin et al., 2021; Machhi et al., 2021; Guerrini et al., 2022)). Apart from the efficacy and immunogenicity of vaccines, safety and protection against reinfections are other challenges that must be considered in the development of vaccines (WHO, 2020; Chen et al., 2021b; Liu et al., 2021c; Hasanzadeh et al., 2021; Teijaro and Farber, 2021). The spike (S) gene/protein of SARS-CoV-2 has been chosen as the main antigen to design vaccines to immunize, either by itself (i.e., in form of mRNA or derived DNA gene or recombinant protein) or present as protein in attenuated or inactive viral particles or in viral like particles (VLPs) (Heinz and Stiasny, 2021; Martinez-Flores et al., 2021) (Figure 3A), because the targeting of this protein with vaccine-elicited nAbs could be important to impair the entry of the virus into the host cells and cause infection (Figure 3B, see inbox: nAbs action in the S-tertiary structure), therefore, inhibiting viral replication, spread and cause the disease (Brouwer et al., 2020; Corbett et al., 2021; Earle et al., 2021; Khoury et al., 2021; McMahan et al., 2021) (see Figure 1A,B).
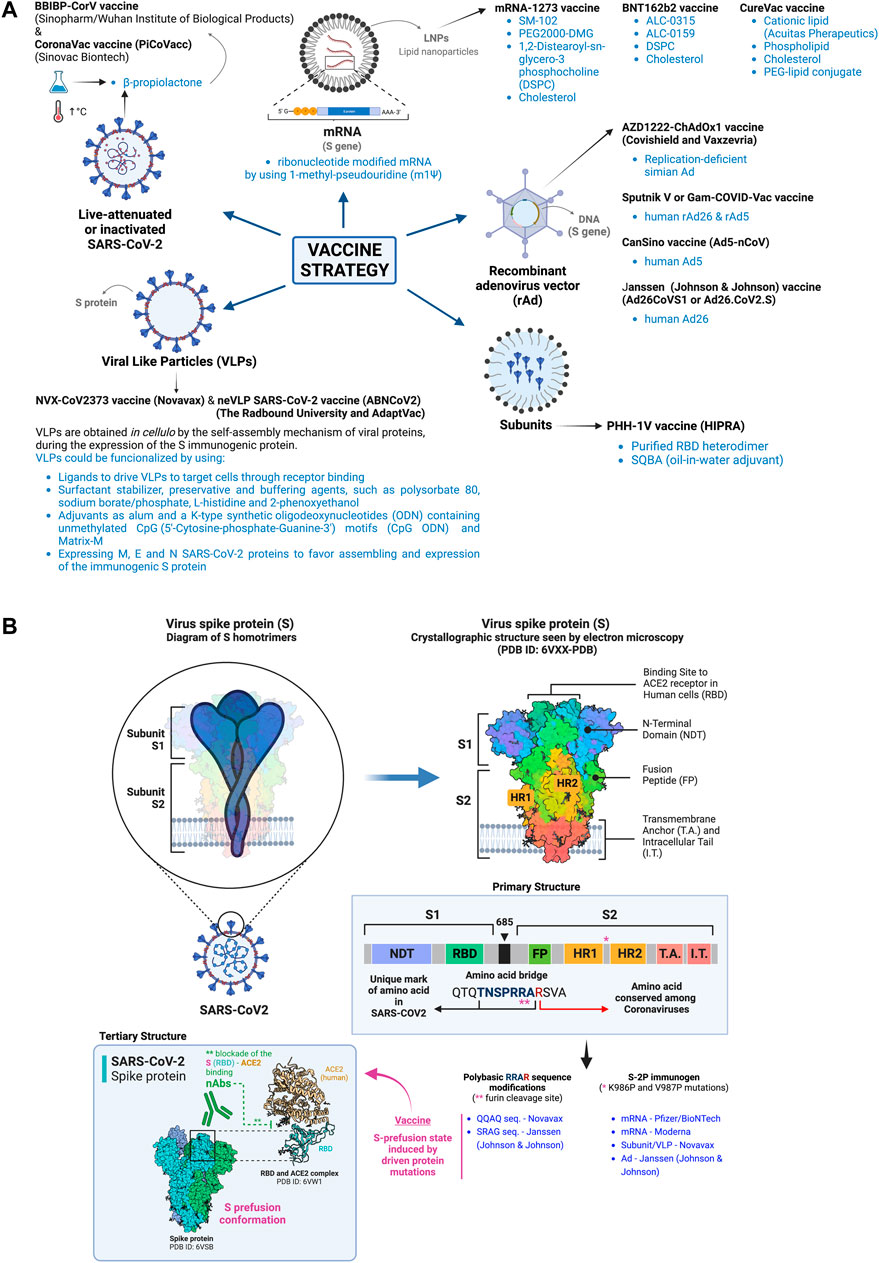
FIGURE 3. Nano-based COVID-19 vaccines. (A) The scheme summarizes the different nano-based strategies used for immunization against SARS-CoV-2 infection and severe COVID-19 based or different nanomaterials and reagents to deliver the modified viral genome (coding for the immunogen S protein) and recombinant immunogen S protein or its subunits, such as the LNP-mRNA vaccines, the rAd vectors containing the DNA S gene, chemically inactivated SARS-CoV-2 viral particles, VLPs bearing recombinant S protein, and recombinant subunit vaccines (i.e., adapted full-length S protein or fragments as the RBD region) in emulsion with nanoreagents. All nanomaterials and nanotechnology used for vaccine development are indicated in the different vaccine strategies. (B) Schemes represent the primary and tertiary structures of the S viral protein, used as immunogen for vaccine developments, stabilized in its prefusion state by two key protein mutations (K986P and V987P; named S-2P immunogen) and modifying the polybasic domain (RRAR) to render it non-cleavable by the furin proteinase. The S protein domains and its trimeric organization on virus membrane and on infected-cell plasma membrane are shown, together with the RBD sequence where nAbs bind to neutralize SARS-CoV-2/ACE2 interaction and subsequent virus infection. The S-associated modifications introduced by the different vaccine designs are indicated. Designs and templates are created with BioRender.
The design and generation of the above indicated genomic or protein-based viral immunogens are part of the nanotechnology employed for vaccination (Chauhan et al., 2020; Wang et al., 2020d), summarized in Figure 3. Moreover, nano-based technology used for vaccine formulation and immunogen administration has been useful to achieve specific anti-viral immune responses and facilitate the positive outcome from severe SARS-CoV-2 infection and illness (Kräutler et al., 2017; Anderson et al., 2020; Walsh et al., 2020a; Walsh et al., 2020b; Chung et al., 2020; Jackson et al., 2020; Mulligan et al., 2020; Polack et al., 2020; Heath et al., 2021; Meo et al., 2021; Turner et al., 2021; Vahedifard and Chakravarthy, 2021; Widge et al., 2021; Toback et al., 2022). Indeed, the use of these NPs or reagents in vaccine designs protect the genomic o protein immunogen from fast degradation by host proteinases, thus enhancing their stability (Miles et al., 2005; Tinkle et al., 2014; Zhao et al., 2014; Chung et al., 2020). Likewise, NPs facilitate the delivery of the vaccine in the organism and the further uptake of viral antigens by antigen-presenting cells (APCs) (Schwendener, 2014; Tinkle et al., 2014; Zhu et al., 2014; Chung et al., 2020). Vaccine stability and solubility has been promoted by the use of NPs such as liposomes (Peek et al., 2008; Alving et al., 2016; Bulbake et al., 2017; Perrie et al., 2017), lipids (Sivakumar et al., 2011; Baz Morelli et al., 2012; Comber and Philip, 2014; Schwendener, 2014; Cibulski et al., 2016), biodegradable polymers (i.e., polyanhydride copolymers based on 1,8-bis-(p-carboxyphenoxy)-3,6-dioxaoctane (CPTEG), 1,6-bis-(p-carboxyphenoxy)-hexane (CPH), and sebacic anhydride (SA)) or dendrimer-based formulations (Chahal et al., 2016; Wafa et al., 2017; Banerjee et al., 2019; Liu et al., 2021b) and emulsions (i.e., MF59®, squalene and Montanide ISA™51) (Miles et al., 2005; Ascarateil and Dupuis, 2006; Della Cioppa et al., 2012; O'Hagan et al., 2013; Della Cioppa et al., 2014; Ascarateil et al., 2015; Shah et al., 2015; van Doorn et al., 2016; Kantipakala et al., 2019; Savoji et al., 2019). The nature or the physicochemical characteristics of these NPs determine the efficiency of antigen delivery and the potential adverse effects observed during immunization (reviewed in (Mitchell et al., 2021)). Thus, lipid-based NPs (LNPs), such as liposomes and emulsions, allow simplicity during NPs formulation and self-assembly, offering high biocompatibility, bioavailability, and flexibility to accommodate different antigen sizes (Fonseca-Santos et al., 2015; Sercombe et al., 2015), since LNPs could be modified for these purposes during synthesis (Alyautdin et al., 2014; Leung et al., 2015a; Sercombe et al., 2015; Sarfraz et al., 2018; Sedighi et al., 2019). Moreover, LNPs allow to deliver hydrophilic, hydrophobic or lipophilic agents, as well as lipophilic and hydrophilic compounds together in the same LNP preparation (Alyautdin et al., 2014; Fonseca-Santos et al., 2015; Sercombe et al., 2015; Sedighi et al., 2019). Although LNPs achieve low encapsulation efficiency, they are the most approved NP system for drug delivery (Fenton et al., 2018; Anselmo and Mitragotri, 2019), as occurred with the mRNA-based vaccines against SARS-CoV-2 infection and COVID-19 disease (reviewed in (Tenchov et al., 2021)). Natural (i.e., heparin, hyaluronan, chitosan or dextran (Lombardo et al., 2019; Zielińska et al., 2020)) or synthetic [i.e., saturated poly (α-hydroxy esters), such as poly (lactic acid) (PLA), poly (glycolic acid) (PGA), and poly (lacticcoglycolide) (PLGA) (Shive and Anderson, 1997; Lin et al., 2012; Chenthamara et al., 2019; Gangapurwala et al., 2020; Zielińska et al., 2020)] polymeric NPs, such as dendrimers or nanospheres, permit the precise control of the physicochemical particle characteristics, allowing posterior surface modifications and being flexible for hydrophilic and hydrophobic cargo accommodation (Kannan et al., 2014; Xu et al., 2014; Patra et al., 2018; Zhang et al., 2020b; Pinelli et al., 2020; Valcourt et al., 2020; Volpatti et al., 2020; Zielińska et al., 2020). These characteristics together with their biocompatibility, biodegradability and low toxicity (Chenthamara et al., 2019; Lombardo et al., 2019; Pinelli et al., 2020) make polymeric NPs a good choice for vaccine delivery (Zhang et al., 2020b). Of note, these types of polymeric NPs could present a high degree of aggregation that could be toxic and difficult antigen delivery, therefore compromising the optimal antigen concentration in target cells and tissues for immunization (Knight et al., 2019; Zielińska et al., 2020).
These NPs or vehicles also favor immunogenicity because they act as adjuvants triggering the immune cell activation and promoting immune responses (Ascarateil and Dupuis, 2006; Sivakumar et al., 2011; Baz Morelli et al., 2012; Zhu et al., 2014; Ascarateil et al., 2015; Varypataki et al., 2015; Alving et al., 2016; Cibulski et al., 2016; Khabazzadeh Tehrani et al., 2016; Perrie et al., 2017; Wafa et al., 2017; Khademi et al., 2018; Kantipakala et al., 2019; Savoji et al., 2019; Petkar et al., 2021). The NP surface could be modified by presenting monosaccharides (i.e., mannose) or immune ligands (i.e., toll-like receptor ligands), in order to improve elicited vaccine delivery of the immunogen and immune stimulation (Apostolopoulos et al., 2013).
In this scenario against SARS-CoV-2/COVID-19, LNPs have been developed and used to deliver and immunize with modified mRNA of the entire S protein or only the RBD segment of the viral glycoprotein (i.e., novel LNP-encapsulated mRNA based vaccine (Wang et al., 2020a) or LNP-encapsulated mRNA encoding RBD vaccine (Tai et al., 2020)) (Figure 3A), or by using a self-amplifying RNA (saRNA or SAM) (Perri et al., 2003) that encodes the alphaviral replicase to drive the expression of a pre-fusion stabilized SARS-CoV-2 S-gene/protein. This strategy allows to reduce the injection dose (tenfold lower than mRNA vaccines) and follow a “one-dose prime-boost” regime (Vogel et al., 2018; Erasmus et al., 2020; Schoenmaker et al., 2021) instead of the “two-dose” strategy per recipient of the non-replicative mRNA-COVID-19 vaccines. SAM vaccines in development are the LNP-nCoV-saRNA vaccine by the Imperial College London (McKay et al., 2020; Schoenmaker et al., 2021), and the ARCT-021 or LUNAR-COV19 vaccine (see information in https://www.nature.com/articles/d43747-021-00073-3) by Arcturus/Duke-NUS (Low et al., 2021; Schoenmaker et al., 2021). The LNP-nCoV-saRNA vaccine has been assayed in a Phase I clinical trial, being immunogenic at low dose levels, but failed to induce 100% seroconversion. Some modifications are being developed to optimize humoral responses against the viral S antigen (Pollock et al., 2022). In the case of ARCT-021 (LUNAR-COV19) vaccine, the mRNA is a replicon that comprises the Venezuelan equine encephalitis virus (VEEV) genome in which the structural genes have been replaced with the SARS-CoV-2 full-length S-gene, and formulated with the proprietary LUNAR® LNP (Low et al., 2021). ARCT-021 protects K-18 human ACE2 transgenic mice from lethal SARS-CoV-2 challenge producing a strong Th1-predominant humoral and cellular immune responses against the S protein (de Alwis et al., 2021), with promising 1–2 phase clinical trial results (Low et al., 2021).
In the case of liposomes, Moderna’s candidate vaccine mRNA-1273 (Moderna/National Institute of Allergy and Infectious Diseases; commercially available under the brand name Spikevax) for COVID-19 used them as a carrier for drug delivering and as an adjuvant, thereby encapsulating the nucleoside modified mRNA that encodes the SARS-CoV-2 S glycoprotein (Callaway, 2020; Huang et al., 2020c; Baden et al., 2021; Dai and Gao, 2021; Tenchov et al., 2021), whereas LNPs were used for this purpose by BioNTech/Fosun Pharma/Pfizer, BNT162b2 mRNA vaccine (commercially available under the brand name Comirnaty) (Callaway, 2020; Polack et al., 2020; Baden et al., 2021; Dai and Gao, 2021; Tenchov et al., 2021) (Figure 3A). The LNPs components of these two efficient mRNA vaccines are: 1) for mRNA-1273, SM-102, PEG2000-DMG, 1,2-Distearoyl-sn-glycero-3 phosphocholine (DSPC) and cholesterol; 2) for BNT162b2, ALC-0315, ALC-0159, DSPC and cholesterol (reviewed in (Kim et al., 2021a; Schoenmaker et al., 2021; Verbeke et al., 2021)) (Figure 3A). Furthermore, Pfizer/BioNTech and Moderna vaccines both uses a ribonucleotide modified mRNA by using 1-methyl-pseudouridine (m1Ψ) instead of uridine (Polack et al., 2020; Baden et al., 2021) (Figure 3A). In fact, the m1Ψ nucleotide is a natural archaeal tRNA component (Wurm et al., 2012) that could be chemically synthesized and used for production of m1Ψ mRNAs by in vitro transcription (reviewed in (Nance and Meier, 2021)). The presence of the m1Ψ nucleotide in the mRNA construct provokes changes in the secondary structure of the mRNA that has been associated with high protein expression (Andries et al., 2015; Zhao and He, 2015; Svitkin et al., 2017; Van der Jeught et al., 2018; Foster et al., 2019; Hadas et al., 2019; Mauger et al., 2019; Nelson et al., 2020; Parr et al., 2020; Nance and Meier, 2021; Mokuda et al., 2022). Likewise, the introduction of these m1Ψ nucleotides inside mRNA fragments has been reported to stabilize the mRNA and reduce its recognition by RNA-binding proteins, thereby minimizing undesirable innate immune responses against this foreign mRNA immunogen (Andries et al., 2015; Zhao and He, 2015; Van der Jeught et al., 2018; Foster et al., 2019; Mauger et al., 2019; Nelson et al., 2020; Nance and Meier, 2021; Mokuda et al., 2022).
Moreover, Pfizer/BioNTech and Moderna vaccines both uses a mRNA encoding a modified S protein stabilized in its prefusion conformation (Erasmus et al., 2020; Hsieh et al., 2020; Wrapp et al., 2020), in order to maintain a stable conformational state exposing the S1 distal domain of the S protein, against which vaccine-elicited protective nAbs are aimed to be generated (Erasmus et al., 2020; Hsieh et al., 2020; Xia, 2021) (Figure 3B, see inbox: nAbs in the S-tertiary structure). The strategy to stabilize the immunogenic prefusion conformation on the membrane-anchored S protein consists in modifying two key amino acid residues for proline in the S protein (i.e. K986P and V987P), leading to the known S-2P immunogen (Walsh et al., 2020b; Corbett et al., 2020; Olia et al., 2021; Xia, 2021) (Figure 3B). These consecutive residues are located in the central helix region of the S protein, a segment placed between the Heptad-Repeat (HR) domains HR1 (912–984 residues) and HR2 (1,163–1,213 residues) of the S protein (Huang et al., 2020c; Hsieh et al., 2020; Xia, 2021) (Figure 3B). The K986P and V987P mutations prevent structural transitions from prefusion to postfusion state, thereby stabilizing the S-2P protein at the prefusion state and contributing to vaccine efficiency (Huang et al., 2020c; Hsieh et al., 2020; Xia, 2021) (Figure 3B). Other vaccine developments that contain the S-2P immunogen are the Sanofi design, the subunit/VLP Novavax vaccine and the Janssen (Johnson and Johnson) adenovirus (Ad)-based vaccine (Guebre-Xabier et al., 2020; Mercado et al., 2020; Goepfert et al., 2021). Furthermore, another strategy to stabilize the immunogenic prefusion conformation of the SARS-CoV-2 S glycoprotein consists in changing the furin cleavage site in the S protein (Andersen et al., 2020), in order to avoid this cleavage that releases the S1 subunit of the S viral protein that would compromise the appropriate immunogenic state of the S protein for vaccination (Walls et al., 2020b; Wrapp et al., 2020; Johnson et al., 2021; Olia et al., 2021). In fact, the SARS-CoV-2 S protein must be cleaved into the S1 and S2 subunits, in order to be able to trigger fusion and host-cell entry (Hoffmann et al., 2020a; Walls et al., 2020b; Jaimes et al., 2020) (Figure 1B). SARS-CoV-2 presents a four-amino acid “SPRRAR|S” sequence (preceded with P) named as the polybasic site or the furin cleavage site, which is inserted at the junction of the S1 (containing the RBD sequence) and S2 (bearing the fusion domain) subunits of the S glycoprotein (Andersen et al., 2020) (Figure 3B). This polybasic site has been associated with the pathogenesis of SARS-CoV-2 infection and COVID-19 pandemic severity (Hoffmann et al., 2020a; Andersen et al., 2020; Shang et al., 2020b; Walls et al., 2020b; Jaimes et al., 2020; Johnson et al., 2020; Lau et al., 2020; Lemmin et al., 2020; Johnson et al., 2021; Whittaker, 2021; Villoutreix et al., 2022). It is interesting to note that the sequence of the ancestral (Wuhan-Hu-1) SARS-CoV-2 furin cleavage site (RRxR) does not follow those of the prototypical furin cleavage sites (RxK/RR) (Tang et al., 2021), and that during the COVID-19 pandemic this site has been adapted in some VOCs, such as B.1.1.7 (Alpha) and B.1.617 (B.1.617.1 named Kappa and B.1.617.2 named Delta) (Aleem et al., 2022) where the first proline has been replaced by a histidine (HRRAR|S) or an arginine (RRRAR|S), respectively (Peacock et al., 2021b; Neerukonda et al., 2021; Whittaker, 2021; Lubinski et al., 2022). Altogether, this information shows that some vaccine designs have incorporated mutations at the furin cleavage site to improve the immunogenic activity of the S-2P immunogen (Figure 3B). Furthermore, the Ad26CoVS1 or Ad26. CoV2 vaccine [Janssen Pharmaceutical Companies (Johnson and Johnson)] presents a substitution of the S-2P “RRAR (R682-R685)” sequence with the non-cleavable SRAG sequence (Bos et al., 2020; Mercado et al., 2020; Dai and Gao, 2021), whereas the NVX-CoV23 vaccine (Novavax) substitutes the furin-polybasic domain (R682-R685) with the non-cleavable QQAQ sequence (Bangaru et al., 2020; Keech et al., 2020; Dai and Gao, 2021; Tian et al., 2021) (Figure 3B).
Noteworthy, other mRNA-based LNP-driven strategies have also been developed, such as the CureVac N.V. vaccine (i.e., CVnCoV vaccine containing sequence-optimized mRNA coding for a stabilized form of SARS-CoV-2 S protein encapsulated in LNP (Kremsner et al., 2021; Rauch et al., 2021; Sáez-Llorens et al., 2022) (Figure 3A), which was expected to be cheaper and more stable in refrigerated storage than those developed by Pfizer-BioNTech and Moderna. These last vaccines contain a pseudouridine mRNA nucleotide in place of the normal uridine nucleotide to circumvent immune inflammatory reactions (Karikó et al., 2005) that could be activated through detection of the foreign S viral mRNA (Morais et al., 2021) (Figure 3A). Although CureVac’s vaccine bears normal uridine (Kremsner et al., 2021), it was developed altering the S viral mRNA genome code in a way that the protein is not affected (Kremsner et al., 2021), but helps to evade immune detection ((Rauch et al., 2021; Sáez-Llorens et al., 2022); analyzed in (Dolgin, 2021)). The formulation of CurveVac’s LNPs consists in cationic lipid (Acuitas Therapeutics), phospholipid, cholesterol and PEG-lipid conjugate (reviewed in (Kim et al., 2021a; Schoenmaker et al., 2021; Verbeke et al., 2021)) (Figure 3A). Unfortunately, CureVac’s mRNA vaccine falls on efficacy in clinical trials (analyzed in (Baker and Dolgin, 2021; Dolgin, 2021)). All the nanocarriers used in the above vaccines protect the immunogen against biodegradation and favor their uptake inside cells that should express and present the immunogen to the immune system (Apostolopoulos et al., 2013; Callaway, 2020; Asdaq et al., 2021; Souri et al., 2022).
Another nano-strategy followed to develop a vaccine against COVID-19 is the use of viral vector vaccines to deliver the S gene and express the S protein of SARS-CoV-2 to trigger specific and protective immune responses (Callaway, 2020; Premkumar et al., 2020; Dai and Gao, 2021), such as well-known adenoviruses (Ads) (Tatsis and Ertl, 2004) (Figure 3A). Hence, recombinant non-replicating Ad (rAd) has been used by the University of Oxford/AstraZeneca (AZD1222-ChAdOx1 vaccine; a replication-deficient simian Ad-vectored vaccine) (commercially available under the brand names Covishield and Vaxzevria), Gamaleya National Research Centre for Epidemiology and Microbiology (Sputnik V or Gam-COVID-Vac; rAd26 and rAd5 vaccines), CanSino Biological Inc./Beijing Institute of Biotechnology (Ad5-nCoV vaccine) and Janssen Pharmaceutical Companies (Johnson and Johnson) (Ad26CoVS1 or Ad26.CoV2.S vaccine) (Zhu et al., 2020a; Bos et al., 2020; Zhu et al., 2020b; Callaway, 2020; Folegatti et al., 2020; Logunov et al., 2020; Mercado et al., 2020; Sadoff et al., 2021a; Sadoff et al., 2021b; Creech et al., 2021; Ewer et al., 2021; Logunov et al., 2021; Ramasamy et al., 2021; Stephenson et al., 2021; Swanson et al., 2021; Voysey et al., 2021) (Figure 3A).
The use of the attenuated or inactivated natural SARS-CoV-2 virus is another strategy that has been applied in this important COVID-19 pandemic crisis (Gao et al., 2020a; Wang et al., 2020b; Xia et al., 2021), due to the potential efficacy and production speed (Figure 3A). This procedure implies the culturing of high amounts of infectious viral strains, isolated from patients, expanded, for example, in infected Vero cells as performed for the BBIBP-CorV (Sinopharm/Wuhan Institute of Biological Products) and CoronaVac vaccine (formerly named PiCoVacc; Sinovac Biontech) vaccines (Gao et al., 2020a; Wang et al., 2020b) (Figure 3A). The Vero continuous cell line (CCL) allows highly efficient virus proliferation and conserves its genetic stability, being both requirements for the development of an inactivated virus vaccine. Hence, high viral genome homology between the expanded virus and the primary strain must be confirmed, as it was analyzed for BBIBP-CorV vaccine with no amino acid variations observed within ten passages (Wang et al., 2020b) or for PiCoVacc vaccine (Gao et al., 2020a). In fact, Vero was the first CCL approved by the WHO to produce human vaccines (WHO, 1987; Barrett et al., 2009; Kiesslich and Kamen, 2020). Thus, amplified and purified infectious viral particles are further inactivated by different procedures. Here, we would like to remark the viral chemical inactivation as performed for the BBIBP-CorV vaccine (Wang et al., 2020b). This is inactivated by using β-propiolactone (at a ratio of 1:4,000 at 2–8°C for 20–24 h) (Figure 3A). The BBIBP-CorV vaccine also contains aluminum hydroxide (alum) as the adjuvant (Wang et al., 2020b) and it has been reported as safe, well tolerated inducing high humoral responses (Xia et al., 2021; Xia et al., 2022). The Sinopharm/Wuhan Institute of Biological Products consortium has also developed two other inactivated virus vaccines based on two primary viral strains, WIV04 and HB02 (Al Kaabi et al., 2021) which offer 72.8 and 78.1% of protection, respectively, against moderate-to-severe forms of the COVID-19 disease. The Sinovac Biontech virus vaccine has been similarly developed (Gao et al., 2020a), expanded in Vero cells and chemically inactivated by using β-propiolactone and mixed with alum adjuvant (Gao et al., 2020a) (Figure 3A). In clinical trials, safety, tolerability and immunogenicity have been also reported for the CoronaVac vaccine (Zhang et al., 2021b; Han et al., 2021; Tanriover et al., 2021; Wu et al., 2021). Bharat Biotech together with the Indian Council of Medical Research and the National Institute of Virology have developed the BBV152 vaccine (Covaxin) (Sharma et al., 2021; Yadav et al., 2021). This inert, safe and quite efficient virus vaccine has been obtained from the NIV-2020-770 isolate, which presents the emerged D614G mutation that increases infectiveness and case fatality rate (Becerra-Flores and Cardozo, 2020; Zhang et al., 2020c; Eaaswarkhanth et al., 2020; Korber et al., 2020; Lorenzo-Redondo et al., 2020; Daniloski et al., 2021; Mansbach et al., 2021; Müller et al., 2021; Plante et al., 2021), and it was formulated with imidazoquinoline (IMDG; a toll-like receptor 7/8 agonist molecule (Miller et al., 2020)) and absorbed in Alum-gel (Algel) as adjuvants (Ella et al., 2021a; Ella et al., 2021b; Sharma et al., 2021; Yadav et al., 2021).
Another nano-based vaccination strategy consist in exposing the immunogenic viral antigen at the surface of empty spherical non-infectious structures named VLPs (viral like particles) (Al-Barwani et al., 2014a; Donaldson et al., 2018), which could be named or classified in enveloped and non-enveloped VLPs (eVLPs and neVLPs, respectively) [reviewed in (Nooraei et al., 2021)] (Figure 3A). VLPs are obtained in cellulo by the self-assembly mechanism of viral proteins, during the expression of the immunogenic protein of interest (Zeltins, 2013; Al-Barwani et al., 2014a). VLP-based vaccines could be considered a kind of subunit vaccines, presenting therefore a characteristic immune response associated with exogenous antigens (Garcea and Gissmann, 2004; Moser et al., 2011; Salvador et al., 2011; Lua et al., 2014; Nooraei et al., 2021). VLPs could be chemically modified by conjugating receptor ligands onto the surface of VLPs to drive their binding to and uptake into APCs expressing the corresponding receptor (Al-Barwani et al., 2014b; Nooraei et al., 2021). Polysorbate 80, sodium borate/phosphate, L-histidine, and 2-phenoxyethanol are often used in the formulation of VLP-based vaccines as a surfactant stabilizer, preservative and buffering agents (Lua et al., 2014) (Figure 3A). VLP-derived vaccines are significantly safer than live-attenuated or inactivated virus vaccines, as well as subunit or particulate vaccines, due to the noninfectious nature of the VLPs (Plummer and Manchester, 2011; Lua et al., 2014; Alon et al., 2017). In fact, some commercially successful vaccines to immunize against human papillomavirus (HPV), hepatitis B virus (HBV) and hepatitis E virus (HEV) are VLP vaccines (Block et al., 2006; Monie et al., 2008; Donaldson et al., 2018; Lee et al., 2019; Qian et al., 2020a). Therefore, VLPs are resembling natural virions but being empty and bearing at the surface membrane of the assembled particle the immunogenic protein (Zeltins, 2013; Al-Barwani et al., 2014a; Donaldson et al., 2018; Mohsen et al., 2020). In the case of COVID-19 vaccine developments, it bears the entire S glycoprotein of SARS-CoV-2 or just the recombinant RBD region of the S protein (Geng et al., 2021; Prates-Syed et al., 2021; Yilmaz et al., 2022). Promising results have been obtained with a VLP-based RBD vaccine which provide highly effective protection against SARS-CoV-2 infection in a mouse model (Geng et al., 2021). Likewise, a VLP-based vaccine carrying an hexaproline prefusion-stabilized S viral protein (S-6p) (Hsieh et al., 2020) has been developed with excellent results in several animal models for SARS-CoV-2 infection (i.e., mice, rats and ferrets) (Yilmaz et al., 2022). This VLP vaccine contains alum and a K-type synthetic oligodeoxynucleotides (ODN) containing unmethylated CpG (5′-Cytosine-phosphate-Guanine-3′) motifs (CpG ODN) as a vaccine adjuvant (Klinman et al., 2009; Ezoe et al., 2020) to promote cellular and humoral immunity (Yilmaz et al., 2022) (Figure 3A). This CpG ODN/alum-adjuvanted 6p-VLP vaccine (VLP-58-1023-Al-K3) is currently being evaluated in a clinical trial (NCT04818281). Furthermore, some VLP developments include other SARS-CoV-2 proteins to stabilize the VLP, such as M (membrane), E (envelope) and N (nucleocapsid) proteins, thereby favoring assembling and expression of the immunogenic S protein (Xu et al., 2020b; Plescia et al., 2021). Worth of mention is the Novavax subunit vaccine (NVX-CoV2373) that uses a recombinant modified nanoparticle S protein of SARS-CoV-2 (Shin et al., 2020b; Chauhan et al., 2020; Keech et al., 2020; Formica et al., 2021; Heath et al., 2021; Tian et al., 2021; Dunkle et al., 2022) together with a Matrix-M adjuvant (Bengtsson et al., 2013) assembled in non-enveloped VLPs (neVLPs) (Bangaru et al., 2020) (Figure 3A). As indicated above, the immunogenic S protein used in this NVX-CoV2373 vaccine is a S-2P protein (1-1273 amino acids) stabilized in the prefusion state, expressed using a baculovirus/insect cell system and then incorporated into polysorbate 80 detergent (Bangaru et al., 2020) (Figure 3B). This full-length S protein has been modified in the polybasic domain at the R682RAR685 sequence, mutated by the Q682QAQ685 sequence (Bangaru et al., 2020; Keech et al., 2020; Tian et al., 2021) (Figure 3B), to avoid its processing by the host furin proteinase (Hoffmann et al., 2020a; Andersen et al., 2020; Shang et al., 2020b; Walls et al., 2020b; Jaimes et al., 2020; Johnson et al., 2020; Lemmin et al., 2020; Johnson et al., 2021; Whittaker, 2021; Villoutreix et al., 2022). The NVX-CoV2373 is a safe vaccine against SARS-CoV-2 infection (Heath et al., 2021), commercially successful as a COVID-19 vaccine (Mahase, 2021; Shinde et al., 2021). The Radbound University and AdaptVac (a company part of the EU H2020 funded PREVENT-nCoV consortium) have also developed a promising neVLP SARS-CoV-2 vaccine (ABNCoV2 vaccine) based on the S RBD (Fougeroux et al., 2021) (Figure 3A), which is currently in phase 1/2 clinical trials (NCT04839146) and phase 3 results are expected by the end of 2022.
On the other hand, recombinant protein-based vaccines are promising candidates for immunization because they offer a safety profile with no live or attenuated viral components in its formulation, avoid potential genomic integration issues, since there are no genomes to be delivered into host cells, and seem to be suitable for immunocompromised individuals (Pollet et al., 2021a; Kleanthous et al., 2021; Kyriakidis et al., 2021) (Figure 3A). The above presented Novavax NVX-CoV2373 vaccine could be also considered as a recombinant protein-based vaccine (Bangaru et al., 2020; Heath et al., 2021; Mahase, 2021; Shinde et al., 2021), as well as some developments focused on the RBD sequence of the S1 subunit of the viral S protein (Malladi et al., 2021b; Kleanthous et al., 2021; Lee et al., 2021) (Figure 3A). The RBD fragment has been assayed as a protein-recombinant vaccine candidate by delivering soluble monomers or dimers of the RBD region (Yang et al., 2020a; Dai et al., 2020; Dalvie et al., 2021a; Malladi et al., 2021a; Malladi et al., 2021b; Pollet et al., 2021b; Chen et al., 2021c; Wang et al., 2021f; Sun et al., 2021), fused to NPs (Walls et al., 2020a; Ma et al., 2020; Arunachalam et al., 2021; Kim et al., 2021b; He et al., 2021; King et al., 2021; Eugenia Toledo-Romaní et al., 2022) or exposed on VLPs (Dalvie et al., 2021a; Dalvie et al., 2021b; Kang et al., 2021; Tan et al., 2021; Zha et al., 2021) (Figure 3A). These recombinant protein-based vaccines present several advantages for development and distribution, since they are produced in large scale, their designs and formulations are safe and immunogenic as those developed using the full-length S protein, they are temperature-stable vaccines and doses have an affordable cost (Malladi et al., 2021b; Kleanthous et al., 2021; Lee et al., 2021). Remarkably, one the most promising recombinant protein vaccines is the new PHH-1V vaccine by HIPRA for intramuscular administration, developed in a consortium with different research centers and Hospitals (Barreiro et al., 2022; Corominas et al., 2022; Leal et al., 2022) (Figure 3A). The PHH-1V vaccine is a highly purified RBD heterodimer (i.e., to avoid limited immunogenicity obtained with monomeric RBD protein (Dai et al., 2020; Yang et al., 2021b)) which is formulated with SQBA, an oil-in-water adjuvant produced by HIPRA, and prepared as an emulsion that does not need to be reconstituted or thawed before administration (Figure 3A). Importantly, the PHH-1V vaccine consists in a fusion heterodimer that contain S-RBD sequences from two SARS-CoV-2 VOCs (Barreiro et al., 2022; Corominas et al., 2022; Leal et al., 2022), B.1.1.7 (Alpha) and B.1.351 (Beta). PHH-1V RBD sequences carry key mutations of epidemic relevant VOCs of SARS-CoV-2, such as the K417N, E484K and N501Y22 humoral escape mutations (Wang et al., 2022a; Wang et al., 2022c), in order to elicit humoral and cellular immune responses against these VOCs or future emerging variants bearing these escape mutations. Hence, the RBD of the S glycoprotein of B.1.351 (Beta), P.1 (Gamma) and B.1.621 (Mu) variants present E484K and N501Y mutations (Wang et al., 2022a; Wang et al., 2022c), whereas both K417N and N501Y immune escape mutations (i.e., for natural and vaccine elicited immunity) are characteristics of the B.1.351 (Beta) and the currently predominant Omicron (B.1.1.529 and BA lineages) variants (Chen et al., 2021a; Wang et al., 2022a). The European Commission has signed a joint procurement contract with the Spanish HIPRA pharmaceutical firm for the supply of the PHH-1V recombinant protein COVID-19 vaccine, which will be commercially available once the European Medicines Agency (EMA) approve it.
COVID-19 vaccines are not protective against SARS-CoV-2 infection and reinfections, due to the absence of antigen specific immunity elicited in the nasal cavities (considered the main route of entry of SARS-CoV-2 into the organism together with oral cavities and ocular conjunctiva) (Cantuti-Castelvetri et al., 2020; Daly et al., 2020; Gengler et al., 2020; Bougakov et al., 2021; Gallo et al., 2021; Huang et al., 2021; Meinhardt et al., 2021; Rodríguez-Ares et al., 2021; Maurin et al., 2022; Pipolo et al., 2022) and the associated mucosal epithelial cells, such as ciliated cells and mucus-producing goblet cells (Lee et al., 2020a; Hou et al., 2020; Osan et al., 2020; Wölfel et al., 2020; Ahn et al., 2021; Bougakov et al., 2021; Meinhardt et al., 2021; Nakayama et al., 2021; Robinot et al., 2021; Morrison et al., 2022; Osan et al., 2022; Pipolo et al., 2022). In these tissues, viral infection is cleared by nasal-associated lymphoid tissue (NALT), including lymphocytes (B and T cells), dendritic cells (DCs), and macrophages. However, these immune responses are not protecting against reinfections (reviewed in (Gallo et al., 2021)). Therefore, sterilizing vaccines should promote mucosal immunity at that level, in order to render nasal cavities and NALT as barriers for SARS-CoV-2 infection and reinfections (reviewed in (Focosi et al., 2022)). In this regard, promising results have been obtained in animal models with the NSP16-deficient SARS-CoV-2 virus, which acts as a live attenuated vaccine that confers sterilizing immunity after intranasal administration of a single dose (Ye et al., 2022). Other studies also reported positive results in animal models (mice, golden hamsters, and ferrets) for nasal-mucosal, inhaled vaccination conferring long-lasting systemic and mucosal immunity (Moreno-Fierros et al., 2020; Mudgal et al., 2020; Ashraf et al., 2021; Park and Lee, 2021; Tiboni et al., 2021; Focosi et al., 2022; Heida et al., 2022). These vaccine developments would achieve the desired sterilizing immunity for mucosal cells and tissues against SARS-CoV-2, just at the door of our organism for viral infection. However, intranasal or mucosal vaccines should follow the tissular dynamics followed by SARS-CoV-2 to accomplish neutralizing immunity, and therefore all designed vaccines and associated adjuvants must be safe, avoiding reaching and affecting the central nervous system (CNS) via the olfactory bulb that could compromise the life of the inoculated individuals. Fortunately, for vaccine developments with SARS-CoV-1 S protein, it has been reported that the use of rAd vectors via sublingual administration induce systemic and mucosal immunity without redirection of the viral rAd vector to the brain (Shim et al., 2012). Similar immunoprotection results have been obtained with a recombinant adeno-associated virus (rAAV) expressing the RBD of the SARS-CoV-1 S protein (RBD-rAAV) intranasal administered (Du et al., 2008). Although these studies show that mucosal immunization against SARS-CoV-1 S protein did not provide full sterilizing immunity, they prevent virus dissemination to the lung and prevent respiratory distress, being appropriated models to achieve SARS-CoV-2 sterilizing vaccines. Moreover, the use of appropriate nanoreagents to form stable and safe nanoparticles for intranasal administration of Abs, proteins or genomic fragments as immunogens would be key to accomplish the desired sterilizing immunity. Thus, a recent study has reported that it is possible to deliver a mRNA to the lung via nebulization of LNPs made of lipids, neutral or cationic helper lipids and polyethylene glycol (PEG). The intranasal administration of nebulized LNPs carrying a mRNA encoding for a broadly nAb targeting haemagglutinin protects mice from lethal challenge of the H1N1 subtype of influenza A virus (Lokugamage et al., 2021).
Therefore, nano-based COVID-19 vaccine designs use different nanomaterials, nanocarriers and technologies to enhance the bioavailability of the viral immunogen, control its delivery and faster uptake by target host cells and tissues, and also acting as adjuvants to activate the immune responses against the viral antigen, in order to promote specific and protective immune activities against SARS-CoV-2 infection and severe COVID-19 disease (reviewed in (Campos et al., 2020; Chauhan et al., 2020; Chung et al., 2020; Talebian and Conde, 2020; Constantin et al., 2021; Creech et al., 2021; Machhi et al., 2021; Rashidzadeh et al., 2021; Varahachalam et al., 2021; Guerrini et al., 2022; Souri et al., 2022)) (Figure 3).
2.3 Virosurveillance: Genomics to trace SARS-CoV-2
Surveillance of infectious diseases, and particularly of emerging and new emerging virus diseases, is key for ensuring community and global health in the XXI century. In 7 years from 2011, WHO alerted and followed nearly 1,500 epidemic events in several countries due to emerging viruses, such as SARS, MERS, influenza, Zika virus (ZIKV), Yellow Fever virus (YFV), and Ebola virus (EBOV), among other viruses (Global-Preparedness-Monitoring-Board-(GPMB), 2019). The world began to realize the fragility of our societies and economies in facing severe non-curable and/or lethal emerging virus when the 2014–2015 ZIKV pandemic seriously affected two of the biggest world sports events: the Olympic Games in Brazil (2016) and the World Soccer Championship in Brazil (2018) (Petersen et al., 2016; Salvador and Fujita, 2016). These epidemic events announce the new global era the world is facing with fast-spreading outbreaks, difficult to manage and having impact on lives, health systems, administration and the economy of companies, families and countries, as is occurring nowadays with the worldwide SARS-CoV-2 infection (Reports, 2020) responsible for the still ongoing COVID-19 pandemic (Beech and Anseel, 2020; Brammer and Clark, 2020; Nicola et al., 2020; Shankar, 2020; Verbeke, 2020; Heinberg and Steffen, 2021; Klebe et al., 2021; Onyeaka et al., 2021; Poudel et al., 2021; Shang et al., 2021; Sheng et al., 2021; Sohrabi et al., 2021; Yen et al., 2021; Chang et al., 2022; Desai et al., 2022). Surveillance of these emerging viruses involves producing information to guide actions at different levels (Buckeridge and Cadieux, 2006) to protect all sectors that are in danger because of the absence of preparedness policies (Global-Preparedness-Monitoring-Board-(GPMB), 2019; Lee et al., 2020c). Therefore, surveillance is a fundamental tool for public health and our societies, where genomics for virus-surveillance has gain relevance as the worldwide tracking system of the different emerged SARS-CoV-2 variants, as it has been demonstrated in different countries. A remarkable example is the first WGS obtained from patients’ viral samples. A few days passed from the first observed Wuhan patients, affected by the new unknown disease, to the world-wide communication of the viral sequence (Ackermann et al., 2020; Cao et al., 2020; Lu et al., 2020).
The knowledge of a complete new CoV genomic sequence (available at GISAID and GenBank (accession number MN908947) (Cao et al., 2020; Lu et al., 2020)), designated as Wuhan-Human 1 CoV (Wuhan-Hu-1 or WHCV) (Ackermann et al., 2020), allowed the rapid development of probes, reagents and tools for specific and sensitive detection of viral sequences in the patients infected by SARS-CoV-2 (Cheng et al., 2020a; Corman et al., 2020; Dutta et al., 2022). One of the first epidemiological parameters that these tools helped to assess was the basic reproduction number or R0 (R naught) value for virus community transmission, estimated from 2.2 to 2.68 for the ancestral Wuhan-Hu-1 strain (Liu et al., 2020e). These tools allow: 1) specific diagnosis and virosurveillance of outbreaks and community virus spreading, 2) to follow how the virus is evolving and the potential relationships between the genome viral sequence, 3) its mutations, virus infectiveness and disease severity, and 4) to understand natural and vaccine-elicited immunity breakthrough events that help to adapt new vaccines and drugs (Alcoba-Florez et al., 2020a; Corman et al., 2020; Ciuffreda et al., 2021a; Alcoba-Florez et al., 2021; Ciuffreda et al., 2022). A historical and paradigmatic genomic virosurveillance global action, near to real-time and an unprecedented scale (Gardy et al., 2015; Lucas et al., 2020a), started after at that moment by fostering a continuous communication and classification of the obtained viral sequences through different resources (e.g., GISAID, GenBank, Nextstrain, Pangolin, among others) (Hadfield et al., 2018; Andersen et al., 2020; Liu et al., 2020d; Khare et al., 2021; O'Toole et al., 2021; Sayers et al., 2021; Underwood et al., 2022)), where WGS, GWAS (genome-wide association study) and nanopore sequencing approaches and technologies have gain relevance for surveillance and diagnose (Arora et al., 2022) (Figure 4).
FIGURE 4. Nanotechnology and reagents used for genome sequencing technologies and serological approaches for SARS-CoV-2 surveillance and diagnose. (A) Viral RNA+ extraction from patients’ samples have allowed to prepare a library or primers for WGS that is very useful for virus surveillance and the detection of VOCs bearing key mutations associated with immune responses escape and vaccine breakthrough. Likewise, viral RNA+ extraction have permitted to develop specific, sensitive, scalable, rapid and low-cost RT-qPCR tests for diagnose, both in hospitals and in PoC facilities. Nanomaterials used for RNA+ isolation and purification that allow scalation of RT-qPCR and other gene-based diagnose tests are indicated. Moreover, nanotechnology and nanoreagents used for serological lateral flow tests permit PoC detection of the viral S and N antigens (i.e., from saliva or anterior nasal swabs (nasopharyngeal exudate or other upper/lower respiratory samples)) or Igs (IgM, IgG and IgA) against SARS-CoV-2 S and N proteins from patient blood samples. All nanomaterials and associated strategies (i.e., lateral flow test, colorimetric test, and RT-qPCR) to detect viral genomes and proteins are shown. (B) SARS-CoV-2 genome sequencing using ONT (Oxford nanopore technology). Rapid sequencing of the SARS-CoV-2 is key for a correct public health response during a pandemic. Through nanopore sequencing, RNA samples are retrotranscribed (1), and the whole genome amplified with various sets of primers (2). Amplified fragments can incorporate barcodes through a transposase (2) and then samples can be pooled and sequenced through nanopores (3). SARS-CoV-2 RNA+ is also read directly by nanopore. Detected current patterns over time define the specific nucleobases in a sequence (3), which can be demultiplexed, assembled and analyzed, thus allowing the detection of sensitive, single point mutations in the viral genome and tracing circulating VOCs (3). Designs and templates are created with BioRender.
WGS provides a deep insight into the DNA sequence of the viral genome and, when combined with metadata analysis at the individual or community level, it has allowed to analyze the origins of the COVID-19 pandemic (Al Kaabi et al., 2021; Singh et al., 2021; Costa et al., 2022), and to characterize outbreaks in a community or region (Alcoba-Florez et al., 2021; Aggarwal et al., 2022; Ciuffreda et al., 2022), hospitals (Rivett et al., 2020), farms (Badiola et al., 2021; Oude Munnink et al., 2021), schools (Huang et al., 2022) and homes (Ladhani et al., 2022), among other social or industrial entities (Bartik et al., 2020; Murti et al., 2021) (Figure 4A). Moreover, public health responses to the COVID-19 pandemic are supported by sequencing WGS from circulating SARS-CoV-2 VOCs and analyzing the risk host factors. WGS enables mapping of community and international viral transmission, thereby identifying and tracking the SARS-CoV-2 emerging variants (Gohl et al., 2020; Rivett et al., 2020; Aggarwal et al., 2022; Obermeyer et al., 2022; Rosenthal et al., 2022) (Figure 4A), even helping in clinical care using samples collected from patients-rapid antigen test devices (Gohl et al., 2020; Arora et al., 2022; Rosenthal et al., 2022). Host genetics is also a key element in this scenario. WGS and genome-wide association studies (GWAS) have been used to identify host genetic factors that predict COVID-19 severity and patient survival. In this regard, it was of special interest the GWAS analysis associating inborn errors of immunity (i.e., mainly INF and TLR genes and associated pathways) with severe COVID-19 (Bastard et al., 2020; Chung et al., 2020; Ellinghaus et al., 2020; Grasselli et al., 2020; The-Covid-Host-Genetics-Initiative, 2020; Al Kaabi et al., 2021; Alcoba-Florez et al., 2021; Bastard et al., 2021; Castano-Jaramillo et al., 2021; Fallerini et al., 2021; Niemi et al., 2021; Pairo-Castineira et al., 2021; Tan et al., 2021; Karlsen, 2022; Loucera et al., 2022; Oh and Shin, 2022). In these critically ill patients, the SARS-CoV-2 infection have generally been diagnosed mainly by specific and sensitive retrotranscription and quantitative polymerase chain reaction (RT-qPCR) technique (COVID-19-Host-Genetics-Initiative, 2021) (Figure 4A). In fact, this quantitative, real-time PCR allows to identify specific features in some concerning variants, like S-gene target failure (SGTF) found in the B.1.1.7 (Alpha) and Omicron variants, further confirmed by S-gene sequencing (Volz et al., 2021; Aggarwal et al., 2022; Wolter et al., 2022). In other studies, single-cell RNA sequencing data has been combined with viral WGS analysis, leading key results corelating the level of expression of CCR1 and CXCR6 chemokine receptors on CD8+ T cells and monocytes with COVID-19 severity (Alsved et al., 2022). The presence of SARS-CoV-2 VOCs are detected by genomic sequence analysis. It was used to identify the first VOCs B.1.1.7 (Alpha), B.1.351 (Beta) and P.1 (Gamma) and those that emerged later on, such as B.1.617.2 (Delta) and Omicron BA.1 and BA.2 lineages associated with enhanced viral transmission and pathogenicity, and immune escape (Akilesh et al., 2021; Alcoba-Florez et al., 2021; Altmann et al., 2021; Hirotsu and Omata, 2021; Nakamichi et al., 2021; Alsafar et al., 2022; Ciuffreda et al., 2022; Harrigan et al., 2022; Lyngse et al., 2022; Wawina-Bokalanga et al., 2022). In fact, the viral WGS is appropriated for the identification of the viral genomic variants that are associated with severe disease patterns (Hoffmann et al., 2021; Hoang et al., 2022; Overton et al., 2022; von Wintersdorff et al., 2022).
One sequencing approach that has gained relevance in the context of real-time surveillance of SARS-CoV-2 entails the long-read sequencing technology from Oxford Nanopore Technologies (ONT) (Corman et al., 2020; Pater et al., 2021). ONT technology relies on a nanoscale modified protein pore, named nanopore, that functions as a biosensor which is embedded in a polymer membrane with electric resistant properties (Deamer et al., 2016; van Dijk et al., 2018) (Figure 4B). Negatively charged single-stranded DNA (ssDNA) or RNA molecules, contained in an electrolytic solution, are driven by an ionic current (created under constant voltage) through the nanopore. These charged nucleic acid molecules move from the negatively charged “cis” side to the positively charged “trans” side of the nanopore (Figure 4B). Importantly, a motor protein that acts as an anchored bar pass the ssDNA or RNA molecules through the nanopore in a controlled manner thereby reducing the translocation speed. RNA or ssDNA changes the ionic current during translocation. Using computational algorithms translate these current changes into the nucleotide sequence present in the sample, allowing real-time sequencing of single molecules (Figure 4B). Moreover, the motor protein presents a helicase activity that unwound the double-stranded DNA (sdDNA) or RNA-DNA duplexes into single-stranded molecules that pass through the nanopore (reviewed in (Deamer et al., 2016; Wang et al., 2021e)). Previously, for assuring SARS-CoV-2 genome sequencing and considering that viral RNA extracted from patients samples could be degraded and not the best quality, a series of shorter amplicons (i.e., 400 nucleotides) are used for amplifying across the virus genome and cover the whole sequence (Figure 4B), thereby the viral RNA is converted into DNA by reverse transcription and PCR is used for amplicons generation to cover all the sequence and identifying overlapping amplicons (Kim et al., 2020a; Bull et al., 2020; Brejová et al., 2021; Stüder et al., 2021; Wang et al., 2022b; Li et al., 2022; Xu et al., 2022) (Figure 4B). Then, samples were barcoded by using specific kits and following ligation with a sequencing adapter, in order to generate a library for nanopore sequencing (Figure 4B). The protocol for nanopore sequencing of tiled PCR-generated amplicon pools has been developed an adapted by the Artic Network (https://artic.network/). These procedures have been used for other emerging RNA viruses and microorganisms (Quick et al., 2016; Quick et al., 2017; Ciuffreda et al., 2021b). ONT or nanopore sequencing offers several advantages to obtain viral WGS because these devices do not require large supporting laboratory infrastructures, are portable and are inexpensive, allowing rapid sequencing analysis to be scalable and at reach for any laboratory (Coloma et al., 2016; Corman et al., 2020; Ciuffreda et al., 2021b). Nanopore sequencing allows to detect single nucleotide variants and small insertion/deletions in circulating SARS-CoV-2 with high accuracy and high sensitivity (Corman et al., 2020; Noh et al., 2020; Baker and Dolgin, 2021; Ciuffreda et al., 2021b; González-Recio et al., 2021; Pater et al., 2021), as for the key mutations in SARS-CoV-2 VOCs (La Rosa et al., 2021) (Figure 4B). Moreover, nanopore technology allows to directly sequence the SARS-CoV-2-RNA + genome from patients’ samples (Viehweger et al., 2019; Kim et al., 2020a; Vacca et al., 2022; Xu et al., 2022) (Figure 4B). Nanopore sequencing combined with a machine learning algorithm permits rapid hospital diagnoses of SARS-CoV-2 from saliva samples of the patients (Horejs, 2021). Nanopore sequencing could be used in SARS-CoV-2 diagnose with other technologies, such as the reverse transcription (RT) loop mediated isothermal amplification (LAMP; (Notomi et al., 2000)) (RT-LAMP; (Huang et al., 2022)) underlying the LamPORE approach (James et al., 2020). LamPORE is considered a high-throughput substitute of RT-qPCRs for rapid and accurate diagnosis of SARS-CoV-2 infection (Peto et al., 2021; Ptasinska et al., 2021) while used for SARS-CoV-2 genotyping (Morsli et al., 2021). Moreover, ONT sequencing has been applied for accurate tracking of intra-hospital SARS-CoV-2 transmission (Løvestad et al., 2021). Thus, this affordable and easy-to-scale viral sequencing technology has been a widespread solution for tracking SARS-CoV-2 VOCs (Rios et al., 2021; Yakovleva et al., 2021; Barbé et al., 2022), and help decision making for local public health agents without the need of large genomic infrastructures.
The ability of SARS-CoV-2 to evolve forces constant virosurveillance and the adaptation of the required tools, especially at the genetic level for tracking emergence of new VOCs (Lucas et al., 2020a). The rapid detection and characterization of new variants is therefore crucial to informing the potential efficacy of vaccines and therapeutics (Lucas et al., 2020a). The next-generation genome sequencing technologies, as well as nanopore allow to rapidly adapt protocols and reagents for accurate and sensitive detection of new emerging SARS-CoV-2 VOCs (Figure 4).
2.4 Point-of-care: Rapid diagnosis and control of outbreaks in the field
Point-of-care (PoC) facilities and detection technologies have been stablished all around the world because they enable decentralized (i.e., from hospitals), rapid, sensitive, low-cost diagnosis of SARS-CoV-2 infection in neighborhoods health offices and identification of COVID-19 symptoms (Kilic et al., 2020; van Dongen et al., 2020; Valera et al., 2021). The PoCs have accomplished a crucial role in epidemiology surveillance and in situ first triage of ill patients, avoiding sample transport to a clinical laboratory for analysis, thereby easing Hospital work and management of more sever COVID-19 patients (Vandenberg et al., 2021). PoCs have been of importance for prevention of SARS-CoV-2 transmission to the community and to determine who requires quarantine. In fact, the above reported, first viral genetic sequences enabled the rapid development of PoC real-time RT-qPCR diagnostic tests specific for the new CoV (Kampf et al., 2020a; Corman et al., 2020; Hoehl et al., 2020; Rothe et al., 2020; Gupta et al., 2021a) (Figure 4A). In these PoCs the technology used for testing patients permits minimum samples processing, the use of saliva or anterior nasal swabs (nasopharyngeal exudate or other upper/lower respiratory samples) for accurate SARS-CoV-2 detection (i.e., viral RNA or mainly S and/or N antigen proteins), as well as Abs able to recognize viral proteins, either in symptomatic or asymptomatic patients (Figure 4B). Therefore, COVID-19 tests are developed for molecular diagnostics (viral genome), antigen (viral proteins) and antibody tests (Kilic et al., 2020; Valera et al., 2021). For personnel using these tests, it is important to know the accurate protocol for taking samples (i.e., it is key to have skills to know the most appropriate moment for taking samples) and use of these techniques, in order to minimize non-specific results (false positives or negatives reads) due to inappropriate handling or inadequate interpretation of the obtained results, since this information is considered together with the symptomatology of patients for making clinical decisions (Jacofsky et al., 2020).
Infected individuals have been determined by using gold-standard COVID-19 diagnosis to detect SARS-CoV-2 genomic fragments in patients’ samples, associated with viral proteins, such as the S glycoprotein, N, E and the RNA-dependent RNA polymerase (RdRp), and using RNA purification and one-step RT-qPCR (Alcoba-Florez et al., 2020a; Alcoba-Florez et al., 2020b; Khailany et al., 2020; Alcoba-Florez et al., 2021) (Figure 4A, see RT-qPCR schemes). As presented above, the Chinese authorities published in GenBank and in the GISAID portal database the complete genome of SARS-CoV-2. Thanks to this contribution, the microbiological diagnosis of COVID-19 has been based on the detection of the viral genetic material (RNA+) of SARS-CoV-2 from the collection of respiratory samples from patients with compatible symptoms (Corman et al., 2020; Dutta et al., 2022). Various laboratories began development for the detection of SARS-CoV-2 in accordance with WHO guidelines, which initially published five protocols for diagnosis using RT-qPCR (Corman et al., 2020; Dutta et al., 2022). The first validated RT-qPCR detection test was developed by the Berlin Institute of Virology (Corman et al., 2020). The laboratory PCR procedure consists in the extraction of nucleic acid and the amplification reaction. These processes could be automated for rapid diagnose and to reduce errors (Cheng et al., 2020a) (Figure 4A, see RT-qPCR schemes). Moreover, this RT-qPCR technique could be adapted to rapidly detect SARS-CoV-2 (i.e. by 1 h), bypassing the time consuming RNA extraction step and accelerating viral detection, without test sensitivity reduction and allowing sample scaling to increase testing capacity by sample pooling, being of importance during severe outbreaks with large number of people to be tested in a short period of time (Alcoba-Florez et al., 2020a; Alcoba-Florez et al., 2020b; Alcoba-Florez et al., 2021). Likewise, functionalized coated magnetic nanoparticles (MNP) with a negatively charged polymer (i.e., created from 3-aminopropyl triethoxysilane (APTES) polymerized with diacrylate-amine to obtain the poly (amino-ester), poly-NH2-MNPs) allows viral RNA extraction and purification, in order to implement the detection of the SARS-CoV-2 virus by RT-qPCR on a large scale (Chacón-Torres et al., 2020) (Figure 4A).
Serological tests are diagnostic methods used for rapid detection of Immunoglobulins (Igs) (i.e., IgG, IgM and IgA) against SARS-CoV-2 S and N proteins (Dinnes et al., 2020; Tuaillon et al., 2020; Yamayoshi et al., 2020). These tests are named “rapid tests” (about 15 min) and consist in immunochromatographic lateral flow assays to detect Abs that indicate that individuals are infected or have been recently infected by SARS-CoV-2 (Figure 4, see lateral flow test schemes). Serological tests for detection of anti-SARS-CoV-2 Abs to support infected individuals diagnose were first used by the Duke-NUS Medical School in Singapore. This test, named cPass™, was developed in a consortium with GenScript Biotech Corporation and A*STAR, and measures nAbs in an hour (Jung et al., 2021). Lateral flow Ab tests for SARS-CoV-2 require a drop of blood from the patient, usually from a finger prick, similar to tests to monitor blood sugar in certain types of diabetes. This capture and binding process produces a color change along the test and control lines that can be seen with the naked eye, producing one, two, or three lines depending on the type of antibodies (Abs) present (IgM or IgG) (Figure 4A, see lateral flow schemes) (Figure 4A). These tests work very differently from the RT-qPCR technique and focus on detecting the patient’s immune response to the virus rather than detecting the virus itself. These Ab tests require knowledge about the structure and key infection epitopes of the viral proteins, specifically, those proteins to which the immune system responds by triggering the production of nAbs that could neutralize the virus (Guo et al., 2020b; Jiang and Zhang, 2020; Zhao et al., 2020). In this matter, the generation of Abs against SARS-CoV-2 begin about the sixth day of the onset of symptoms, when a decrease in viral load could be observed. Half of the infected cases have total Abs around the seventh day post-infection and 15 days after infection, almost 100% of infected individuals presented Abs both in mild and severe cases (Guo et al., 2020b; Jacofsky et al., 2020; Long et al., 2020; Zhao et al., 2020). These Ab-based techniques seek to detect the immune response of patients, which increases as the infection progresses, and therefore offer the possibility to detect active disease (Jacofsky et al., 2020). Of note, the presence of Abs does not exclude the possibility of continuing to be contagious and is not indicative of having sterilized immunity against recurrent SARS-CoV-2 infections, either in vaccinated or in vaccinated people where reinfections frequently occur (Bergwerk et al., 2021; Gupta et al., 2021b; Hagan et al., 2021; Song et al., 2021; To et al., 2021; Acharya et al., 2022; Kuhlmann et al., 2022; Ren et al., 2022; Singanayagam et al., 2022).
Antigen serological tests are based on the detection of viral proteins specific to SARS-CoV-2, such as the N protein and the S1 or S2 subunits of the S viral glycoprotein (reviewed in (Falzone et al., 2021; Li and Li, 2021)) (Figure 4A, see lateral flow tests for immune assays). Considering that viral replication is more accentuated in the acute phase, the antigenic test should be carried out in the first five-seven days after the onset of symptoms. The biological samples used to detect viral proteins come from nasopharyngeal, oropharyngeal or sputum exudates (reviewed in (Falzone et al., 2021; Li and Li, 2021)). The test contains a strip of nitrocellulose paper where a line has been painted with Abs that adhere to the virus (Figure 4, see lateral flow test schemes). The sample is mixed with a diluent to be dispensed into the test in the window intended for the sample. In this sample-window, the sample is met by colloidal gold- or latex-labeled Abs that make them visible. Later, in the test line (T), more Abs (unmarked) are placed that will serve to confirm whether or not there is an infection. It seems that the viral load is higher in sputum and nasopharynx, being higher in the initial stages of infection, but if the sample viral load is in the limit of detection, false negative reads will occur, instead the person is infected (reviewed in (Falzone et al., 2021)).
Innovative tools for SARS-CoV-2 detection have also been developed using nanomaterials and derived technology. Multiple characteristics have made these nanomaterials good candidates for diagnosis support: the variability in size, structure and geometry, the surface reactivity that have allowed their functionalization with other biomolecules, the electrical conductivity and/or magnetic properties or the quantum or fluorescent characteristics (Kang et al., 2009; Ansari et al., 2010; Cao et al., 2011; Holzinger et al., 2014; Sun et al., 2017; Mitchell et al., 2021). For example, gold nanoparticles (AuNPs), magnetic NPs and quantum dots (QDs) have been engineered to detect SARS-CoV-2 genes and proteins, as well as anti-SARS-CoV-2 Igs by using optical, fluorescence, magnetic and optomagnetic biosensors (reviewed in (Moabelo et al., 2021)) (Figure 4A, lateral flow tests and colorimetric assays). Thus, AuNPs have been used in colorimetric assays for detecting target regions on N and RdRp viral genes (Moitra et al., 2020; Kumar et al., 2022) (Figure 4A), or by using quantitative paper-based electrochemical sensor chip to detect the N gene (Alafeef et al., 2020). Moreover, AuNPs allow to detect specific IgM and IgG Abs that recognizes SARS-CoV-2 proteins (S and N) by using a colloidal AuNPs-based lateral-flow assay, where AuNPs are coated with S or N viral proteins to capture Abs (Liu et al., 2020a; Huang et al., 2020b; Peng et al., 2021b) (Figure 4A, lateral flow tests for viral antigen detection). AuNPs-based lateral flow tests allow to detect the S or N viral proteins by using S or N protein monoclonal Abs (mAbs) coated to the AuNPs (Mertens et al., 2020; Li et al., 2021a; Liu et al., 2021a), or with sialic acid-coated AuNPs for S protein detection (Baker et al., 2020) (Figure 4A). Likewise, gold nanoislands (AuNIs) have been useful to detect viral genes, such as ORF1ab, RdRp and E by using functional plasmonic photothermal biosensors (Qiu et al., 2020). A real-time optomagnetic biosensor has been applied to detect the RdRp gene by using iron oxide NPs (IONPs) (Tian et al., 2020), whereas graphene oxide (GO) nanosheets, coupled to an anti-S protein Ab, have been used to detect the S antigen in clinical samples by a field-effect transistor (FET)-based biosensing device (Seo et al., 2020). The fluorescence-linked immunoassay technique allows to detect anti-SARS-CoV-2-specific IgG Abs from serum by using MnFe3O4 nanospheres and quantum dot (QD) nanobeads (Guo et al., 2020a). Noteworthy, dye streptavidin coated polymer NPs (SA-DNPs) have been used to detect ORF1ab and N genes by using a multiplex reverse transcription LAMP (mRT-LAMP) coupled with a nanoparticle-based lateral flow biosensor, termed a mRT-LAMP-LFB assay (Zhu et al., 2020d).
These tests are generally low-cost methods useful for PoC facilities and of importance in low-resource settings and regions (De Nardo et al., 2020; Fauziah et al., 2021). Therefore, these genomic, Abs and antigen rapid tests have demonstrated their crucial application for virus surveillance, detection and control of viral outbreaks and community viral transmissions, assisting health systems to rapidly derive only severe ill patients to Hospitals (De Nardo et al., 2020; Mardian et al., 2021) (Figure 4).
3 Concluding remarks
The COVID-19 pandemic has demonstrated the relevance of nanomaterials, reagents and technology to combat SARS-CoV-2 infection, community outbreaks and international spreading of the virus and disease. The use of nanotechnology has made possible to detect virus genomic and protein materials from patients’ samples in a rapid, specifically and affordable manner, being able to assume thousands of samples to be tested per day at microbiology hospital departments. This is a task of great magnitude never faced before at region and international levels. Moreover, nanomaterials and derived diagnose techniques (i.e., RT-qPCR and serology test) are responsible for the success of PoC facilities to undertake the mission of virus outbreak control in cities, towns and neighborhoods, and protecting health worker for infection by wearing high-protective PPEs. PoCs have assured rapid and accurate diagnose of infected individuals, helping to derive sever ill patients to the hospital, and assenting public health policies and administrations to make epidemiological and economic decisions.
Although several drugs and Abs have been used for COVID-19 treatment, there are no specific anti-viral treatments available to eradicate SARS-CoV-2 from the organism and revert severe COVID-19 clinical condition in terminal patients (Drożdżal et al., 2021). For this reason, we have decided not to address all the nano-based drug developments and related delivery systems. However, several nanomaterials and technology are candidate to help in the prevention, diagnosis and treatment of SARS-CoV-2 infection and COVID-19 disease. This is the case of some RNA interference (RNAi) formulations to knock-down key viral genes to limit or inhibit viral infection and progression (Idris et al., 2021; Zhang et al., 2022b; Saify Nabiabad et al., 2022). Several in vitro studies using these RNAi preparations have obtained promising results against SARS-CoV-2. However, the in vivo efficacy of these nanoreagents have been compromised or not yet explored as in clinical trials. Likewise, carbon-based nanomaterials (CBNs) have been developed to analyze their effect on SARS-CoV-2, such as graphene, fullerene, or carbon dots that have been positively assayed on other RNA+ viruses in vitro (reviewed in (Serrano-Aroca et al., 2021)). It seems that graphene derivatives with long aliphatic chains limit SARS-CoV-2 replication with some extension, without observing significant cellular toxicity (Donskyi et al., 2021). These CBNs have been tested in other RNA+ viruses with biocompatibility and capacity to induce tissue regeneration, thereby representing promising nanotools for SARS-CoV-2 treatment. Moreover, and considering the antimicrobial effects reported for silver nanoparticles (AgNPs), thought to be associated with the positive charge of silver ions (Ag+), some works have explored the inhibition of SARS-CoV-2 infection with AgNPs (Jeremiah et al., 2020). These silver, metal nanoparticles have been assayed in clinical trial as Argovit® (https://clinicaltrials.gov/ct2/show/NCT04894409). The iron oxide nanoparticles (IONPs) Fe3O4 and Fe2O3 have been reported to bind the S viral protein of SARS-CoV-2 (Abo-Zeid et al., 2020). Although there are not clinical studies initiated with these metal IONPs, molecular docking studies point to this possibility. Another metal nanoparticle made of Au-NP, ZnO-NP and ClO2 (named TPNT1) has been assayed against SARS-CoV-2. TPNT1 blocks the SARS-CoV-2 viral entry in cells by inhibiting the binding of the S viral protein to the ACE2 receptor (Chang et al., 2021). Noteworthy, the poly (lactic-co-glycolic acid) (PLGA) forms biodegradable polymeric nanoparticles that have been used to deliver several antiviral drugs to treat ill patients suffering the COVID-19 disease, such as Ivermectin (i.e., Ivermectin-loaded PLGA-b-PEG-MAL nanoparticles). PLGA-derived nanoparticles allow to deliver a more potent antiviral dose of the drug and also impairs the S protein/ACE2 binding (Surnar et al., 2020). Several bionanoparticles made of polysaccharides and proteins (i.e., chitosan, alginate, gelatin, and albumin), have been tested or are developed to assay their anti-SARS-CoV-2 activity (reviewed in (Debnath and Srivastava, 2022)). The lack or low toxicity and the excellent biocompatibility showed by all the above-described nanomaterials provide great potential in the development of safe therapeutics based on nanoreagents and technology to prevent SARS-CoV-2 infection and cure the COVID-19 disease.
The use of nanomaterials and technology has reached the major relevance to combat COVID-19 pandemic with the development of nano-based vaccines. In this review, we have addressed the main vaccine designs, prototypes developments and commercial vaccines that have been used to immunize the whole population, in order to protect either individuals at risk or the rest of people. Vaccines have helped to reduce casualties and hospital distress during the last COVID-19 wave. However, these vaccines are not eliciting sterilizing immunity, and vaccinated people could be infected a reinfected by SARS-CoV-2, by the same variant and new evolving emerging variants. The virus could be transmitted between vaccinated people and naturally immunized individuals. Therefore, sterilizing vaccines to immunize at the mucosal, nasal tissues and NALT are a great challenge that nano-based vaccines would be able to achieve in a near future.
Nanotechnology and reagents based on next generation technologies for virus genomic sequencing have similarly achieved a historical milestone, allowing virus surveillance in real time, determining SARS-CoV-2 VOCs and identifying key viral mutations that confer virus resistance against natural- and vaccine-elicited cellular and humoral responses immune escape. This information has been key for making public health decisions for advising vaccine booster shots and wearing masks in public transport, plains, hospitals, schools, bars, companies, and administrations. These genomic sequencing technologies have also been useful in PoC facilities, as nanopore-based approaches have efficiently accomplished, even in low-resources regions. The rapid detection and characterization of new variants is therefore crucial to informing the potential efficacy of vaccines and drugs/treatments.
Altogether, nano-based vaccines and rapid, specific tests for diagnose and outbreak control would help to the efficient control of future COVID-19 epidemic or pandemic episodes. These COVID-19 tools, for sure, will help to the fast development of new and specific nanotools and treatments to challenge future emerging virus and associated crisis that would occur in this XXI century, associated with our global style of life, global warming, and the unsustainable manner we must exploit the resources and biodiversity of our planet.
Author contributions
AV-F wrote the paper, conceived, designed the schemes, supervised and edited the manuscript, and equally contributed to this manuscript with conceptualization and literature research with JG-L and CF, RC-R and SP-Y designed all figures supervised and edited the manuscript. All authors have read and supervised the manuscript and agreed to the published version of the manuscript.
Funding
This work was funded by the European Regional Development Fund (ERDF), PID2021-123031OB-I00 (“Ministerio de Ciencia e Innovación”, Spain), ProID2020010093 (“Agencia Canaria de Investigación, Innovación y Sociedad de la Información” and the European Social Fund), UNLL10-3E-783 (ERDF and “Fundación CajaCa-narias”), “SEGAI-ULL”, and EU funding from the European Centre for Disease Prevention and Control (ECDC/HERA/2021/024 ECD.12241). SP-Y is funded by “Contrato Predoctoral Ministerio-ULL Formación de Doctores” (2019 Program) (“Ministerio de Ciencia, Innovación y Universidades”, Spain). RC-R is funded by RD16/0025/0011 and ProID2020010093 (“Agencia Canaria de Investigación, Innovación y Sociedad de la Información” and European Social Fund). JG-L is funded by the “Juan de la Cierva de Incorporación” Spanish Program (IJC2019-038902-I) (“Ayudas Juan de la Cierva de incorporación; Agencia Estatal de Investigación. Ministerio de Ciencia e Innovación”).
Acknowledgments
We would like to thank BioRender (https://app.biorender.com/) for offering online all outstanding designs and templates thar have allow to design all figures. We have the BioRender licensing rights for all figures.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abo-Zeid, Y., Ismail, N. S. M., Mclean, G. R., and Hamdy, N. M. (2020). A molecular docking study repurposes FDA approved iron oxide nanoparticles to treat and control COVID-19 infection. Eur. J. Pharm. Sci. 153, 105465. doi:10.1016/j.ejps.2020.105465
Abraham, A. M., Kannangai, R., and Sridharan, G. (2008). Nanotechnology: A new frontier in virus detection in clinical practice. Indian J. Med. Microbiol. 26, 297–301. doi:10.4103/0255-0857.43551
Acharya, C. B., Schrom, J., Mitchell, A. M., Coil, D. A., Marquez, C., Rojas, S., et al. (2022). Viral load among vaccinated and unvaccinated, asymptomatic and symptomatic persons infected with the SARS-CoV-2 Delta variant. Open Forum Infect. Dis. 9, ofac135. doi:10.1093/ofid/ofac135
Ackermann, M., Verleden, S. E., Kuehnel, M., Haverich, A., Welte, T., Laenger, F., et al. (2020). Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in covid-19. N. Engl. J. Med. Overseas. Ed. 383, 120–128. doi:10.1056/nejmoa2015432
Aggarwal, D., Myers, R., Hamilton, W. L., Bharucha, T., Tumelty, N. M., Brown, C. S., et al. (2022). The role of viral genomics in understanding COVID-19 outbreaks in long-term care facilities. Lancet Microbe 3, e151–e158. doi:10.1016/s2666-5247(21)00208-1
Ahmed, W., Angel, N., Edson, J., Bibby, K., Bivins, A., O'brien, J. W., et al. (2020). First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 728, 138764. doi:10.1016/j.scitotenv.2020.138764
Ahn, J. H., Kim, J., Hong, S. P., Choi, S. Y., Yang, M. J., Ju, Y. S., et al. (2021). Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19. J. Clin. Invest. 131, 148517. doi:10.1172/jci148517
Akhmetzhanov, A. R., Jung, S. M., Cheng, H. Y., and Thompson, R. N. (2021). A hospital-related outbreak of SARS-CoV-2 associated with variant epsilon (B.1.429) in taiwan: Transmission potential and outbreak containment under intensified contact tracing, january-february 2021. Int. J. Infect. Dis. 110, 15–20. doi:10.1016/j.ijid.2021.06.028
Akilesh, S., Nicosia, R. F., Alpers, C. E., Tretiakova, M., Hsiang, T. Y., Gale, M., et al. (2021). Characterizing viral infection by electron microscopy: Lessons from the coronavirus disease 2019 pandemic. Am. J. Pathol. 191, 222–227. doi:10.1016/j.ajpath.2020.11.003
Al Kaabi, N., Zhang, Y., Xia, S., Yang, Y., Al Qahtani, M. M., Abdulrazzaq, N., et al. (2021). Effect of 2 inactivated SARS-CoV-2 vaccines on symptomatic COVID-19 infection in adults: A randomized clinical trial. Jama 326, 35–45. doi:10.1001/jama.2021.8565
Alafeef, M., Dighe, K., Moitra, P., and Pan, D. (2020). Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano 14, 17028–17045. doi:10.1021/acsnano.0c06392
Al-Barwani, F., Donaldson, B., Pelham, S. J., Young, S. L., and Ward, V. K. (2014a). Antigen delivery by virus-like particles for immunotherapeutic vaccination. Ther. Deliv. 5, 1223–1240. doi:10.4155/tde.14.74
Al-Barwani, F., Young, S. L., Baird, M. A., Larsen, D. S., and Ward, V. K. (2014b). Mannosylation of virus-like particles enhances internalization by antigen presenting cells. Plos One 9, e104523. doi:10.1371/journal.pone.0104523
Alcoba-Florez, J., Gil-Campesino, H., Artola, D. G., González-Montelongo, R., Valenzuela-Fernández, A., Ciuffreda, L., et al. (2020a). Sensitivity of different RT-qPCR solutions for SARS-CoV-2 detection. Int. J. Infect. Dis. 99, 190–192. doi:10.1016/j.ijid.2020.07.058
Alcoba-Florez, J., González-Montelongo, R., Íñigo-Campos, A., De Artola, D. G., Gil-Campesino, H., Ciuffreda, L., et al. (2020b). Fast SARS-CoV-2 detection by RT-qPCR in preheated nasopharyngeal swab samples. Int. J. Infect. Dis. 97, 66–68. doi:10.1016/j.ijid.2020.05.099
Alcoba-Florez, J., Gil-Campesino, H., García-Martínez De Artola, D., Díez-Gil, O., Valenzuela-Fernández, A., González-Montelongo, R., et al. (2021). Increasing SARS-CoV-2 RT-qPCR testing capacity by sample pooling. Int. J. Infect. Dis. 103, 19–22. doi:10.1016/j.ijid.2020.11.155
Aleem, A., Akbar Samad, A. B., and Slenker, A. K. (2022). “Emerging variants of SARS-CoV-2 and novel therapeutics against coronavirus (COVID-19),” in StatPearls (Treasure Island (FL): StatPearls Publishing Copyright © 2022, StatPearls Publishing LLC.).
Alejo-Cancho, I., Gual-De-Torrella, A., Gallego, M., Urrutikoetxea-Gutierrez, M., Lejarraga, C., and López De Goikoetxea, M. J. (2022). Misidentification of the SARS-CoV-2 Mu variant using commercial mutation screening assays. Arch. Virol. 167, 1141–1144. doi:10.1007/s00705-022-05395-w
Allen, C. D., Okada, T., and Cyster, J. G. (2007). Germinal-center organization and cellular dynamics. Immunity 27, 190–202. doi:10.1016/j.immuni.2007.07.009
Alon, D., Stein, G. Y., Hadas-Golan, V., Tau, L., Brosh, T., and Turner, D. (2017). Immunogenicity of sci-B-vac (a third-generation hepatitis B vaccine) in HIV-positive adults. Isr. Med. Assoc. J. 19, 143–146.
Alsafar, H., Albreiki, M., Mousa, M., Azman, S. K., Vurivi, H., Waasia, F., et al. (2022). Genomic epidemiology and emergence of SARS-CoV-2 variants of concern in the United Arab Emirates. Sci. Rep. 12, 14669. doi:10.1038/s41598-022-16967-w
Al-Sharif, E., Strianese, D., Almadhi, N. H., D'aponte, A., Dell'omo, R., Di Benedetto, R., et al. (2021). Ocular tropism of coronavirus (CoVs): A comparison of the interaction between the animal-to-human transmitted coronaviruses (SARS-CoV-1, SARS-CoV-2, MERS-CoV, CoV-229E, NL63, OC43, HKU1) and the eye. Int. Ophthalmol. 41, 349–362. doi:10.1007/s10792-020-01575-2
Alsved, M., Nygren, D., Thuresson, S., Medstrand, P., Fraenkel, C. J., and Londahl, J. (2022). SARS-CoV-2 in exhaled aerosol particles from Covid-19 cases and its association to household transmission. Clin. Infect. Dis. 75, e50–e56. doi:10.1093/cid/ciac202
Altmann, D. M., Boyton, R. J., and Beale, R. (2021). Immunity to SARS-CoV-2 variants of concern. Science 371, 1103–1104. doi:10.1126/science.abg7404
Alving, C. R., Beck, Z., Matyas, G. R., and Rao, M. (2016). Liposomal adjuvants for human vaccines. Expert Opin. Drug Deliv. 13, 807–816. doi:10.1517/17425247.2016.1151871
Alyautdin, R., Khalin, I., Nafeeza, M. I., Haron, M. H., and Kuznetsov, D. (2014). Nanoscale drug delivery systems and the blood-brain barrier. Int. J. Nanomedicine 9, 795–811. doi:10.2147/IJN.S52236
Amaechi, U. A., Sodipo, B. O., Nnaji, C. A., Owoyemi, A., Omitiran, K., Okedo-Alex, I. N., et al. (2020). Social approaches to COVID-19 pandemic response: Effectiveness and practicality in sub-saharan africa. Pan Afr. Med. J. 37, 2. doi:10.11604/pamj.supp.2020.37.2.25183
Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C., and Garry, R. F. (2020). The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452. doi:10.1038/s41591-020-0820-9
Anderson, E. J., Rouphael, N. G., Widge, A. T., Jackson, L. A., Roberts, P. C., Makhene, M., et al. (2020). Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. Overseas. Ed. 383, 2427–2438. doi:10.1056/nejmoa2028436
Andries, O., Mc Cafferty, S., De Smedt, S. C., Weiss, R., Sanders, N. N., and Kitada, T. (2015). N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 217, 337–344. doi:10.1016/j.jconrel.2015.08.051
Anfinrud, P., Bax, C. E., Stadnytskyi, V., and Bax, A. (2020). Could SARS-CoV-2 be transmitted via speech droplets? medRxiv. doi:10.1101/2020.04.02.20051177
Ansari, A. A., Alhoshan, M., Alsalhi, M. S., and Aldwayyan, A. S. (2010). Prospects of nanotechnology in clinical immunodiagnostics. Sensors (Basel) 10, 6535–6581. doi:10.3390/s100706535
Anselmo, A. C., and Mitragotri, S. (2019). Nanoparticles in the clinic: An update. Bioeng. Transl. Med. 4, e10143. doi:10.1002/btm2.10143
Apostolopoulos, V., Thalhammer, T., Tzakos, A. G., and Stojanovska, L. (2013). Targeting antigens to dendritic cell receptors for vaccine development. J. Drug Deliv. 2013, 1–22. doi:10.1155/2013/869718
Arons, M. M., Hatfield, K. M., Reddy, S. C., Kimball, A., James, A., Jacobs, J. R., et al. (2020). Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. Overseas. Ed. 382, 2081–2090. doi:10.1056/nejmoa2008457
Arora, P., Rocha, C., Kempf, A., Nehlmeier, I., Graichen, L., Winkler, M. S., et al. (2021). The spike protein of SARS-CoV-2 variant A.30 is heavily mutated and evades vaccine-induced antibodies with high efficiency. Cell. Mol. Immunol. 18, 2673–2675. doi:10.1038/s41423-021-00779-5
Arora, P., Kempf, A., Nehlmeier, I., Graichen, L., Winkler, M. S., Lier, M., et al. (2022). SARS-CoV-2 variants C.1.2 and B.1.621 (Mu) partially evade neutralization by antibodies elicited upon infection or vaccination. Cell Rep. 39, 110754. doi:10.1016/j.celrep.2022.110754
Arslan, M., Xu, B., and Gamal El-Din, M. (2020). Transmission of SARS-CoV-2 via fecal-oral and aerosols-borne routes: Environmental dynamics and implications for wastewater management in underprivileged societies. Sci. Total Environ. 743, 140709. doi:10.1016/j.scitotenv.2020.140709
Arunachalam, P. S., Walls, A. C., Golden, N., Atyeo, C., Fischinger, S., Li, C., et al. (2021). Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature 594, 253–258. doi:10.1038/s41586-021-03530-2
Ascarateil, S., and Dupuis, L. (2006). Surfactants in vaccine adjuvants: Description and perspectives. Vaccine 24 (2), S83–S8385. doi:10.1016/j.vaccine.2005.01.134
Ascarateil, S., Puget, A., and Koziol, M.-E. (2015). Safety data of Montanide ISA 51 VG and Montanide ISA 720 VG, two adjuvants dedicated to human therapeutic vaccines. J. Immunother. Cancer 3, P428. doi:10.1186/2051-1426-3-s2-p428
Asdaq, S. M. B., Ikbal, A. M. A., Sahu, R. K., Bhattacharjee, B., Paul, T., Deka, B., et al. (2021). Nanotechnology integration for SARS-CoV-2 diagnosis and treatment: An approach to preventing pandemic. Nanomater. (Basel) 11, 1841. doi:10.3390/nano11071841
Ashraf, M. U., Kim, Y., Kumar, S., Seo, D., Ashraf, M., and Bae, Y. S. (2021). COVID-19 vaccines (revisited) and oral-mucosal vector system as a potential vaccine platform. Vaccines (Basel) 9, 171. doi:10.3390/vaccines9020171
Aydillo, T., Gonzalez-Reiche, A. S., Aslam, S., Van De Guchte, A., Khan, Z., Obla, A., et al. (2020). Shedding of viable SARS-CoV-2 after immunosuppressive therapy for cancer. N. Engl. J. Med. Overseas. Ed. 383, 2586–2588. doi:10.1056/nejmc2031670
Baden, L. R., El Sahly, H. M., Essink, B., Kotloff, K., Frey, S., Novak, R., et al. (2021). Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. Overseas. Ed. 384, 403–416. doi:10.1056/nejmoa2035389
Badiola, J. J., Otero, A., Sevilla, E., Marín, B., García Martínez, M., Betancor, M., et al. (2021). SARS-CoV-2 outbreak on a Spanish mink farm: Epidemiological, molecular, and pathological studies. Front. Vet. Sci. 8, 805004. doi:10.3389/fvets.2021.805004
Bae, S. H., Shin, H., Koo, H. Y., Lee, S. W., Yang, J. M., and Yon, D. K. (2020). Asymptomatic transmission of SARS-CoV-2 on evacuation flight. Emerg. Infect. Dis. 26, 2705–2708. doi:10.3201/eid2611.203353
Bae, S., Kim, J. Y., Lim, S. Y., Park, H., Cha, H. H., Kwon, J. S., et al. (2021). Dynamics of viral shedding and symptoms in patients with asymptomatic or mild COVID-19. Viruses 13, 2133. doi:10.3390/v13112133
Baig, A. M., Khaleeq, A., Ali, U., and Syeda, H. (2020). Evidence of the COVID-19 virus targeting the CNS: Tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS Chem. Neurosci. 11, 995–998. doi:10.1021/acschemneuro.0c00122
Baker, N., and Dolgin, E. (2021). Coronapod: CureVac disappoints in COVID vaccine trial. Nature. doi:10.1038/d41586-021-01694-5
Baker, A. N., Richards, S. J., Guy, C. S., Congdon, T. R., Hasan, M., Zwetsloot, A. J., et al. (2020). The SARS-COV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent. Sci. 6, 2046–2052. doi:10.1021/acscentsci.0c00855
Bal, A., Destras, G., Gaymard, A., Stefic, K., Marlet, J., Eymieux, S., et al. (2021). Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020. Eurosurveillance 26. doi:10.2807/1560-7917.es.2021.26.3.2100008
Balagna, C., Perero, S., Percivalle, E., Nepita, E. V., and Ferraris, M. (2020). Virucidal effect against coronavirus SARS-CoV-2 of a silver nanocluster/silica composite sputtered coating. Open Ceram. 1, 100006. doi:10.1016/j.oceram.2020.100006
Banerjee, K., Gautam, S. K., Kshirsagar, P., Ross, K. A., Spagnol, G., Sorgen, P., et al. (2019). Amphiphilic polyanhydride-based recombinant MUC4β-nanovaccine activates dendritic cells. Genes Cancer 10, 52–62. doi:10.18632/genesandcancer.189
Bangaru, S., Ozorowski, G., Turner, H. L., Antanasijevic, A., Huang, D., Wang, X., et al. (2020). Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Science 370, 1089–1094. doi:10.1126/science.abe1502
Bangash, M. N., Patel, J., and Parekh, D. (2020). COVID-19 and the liver: Little cause for concern. Lancet Gastroenterol. Hepatol. 5, 529–530. doi:10.1016/s2468-1253(20)30084-4
Barbé, L., Schaeffer, J., Besnard, A., Jousse, S., Wurtzer, S., Moulin, L., et al. (2022). SARS-CoV-2 whole-genome sequencing using Oxford nanopore technology for variant monitoring in wastewaters. Front. Microbiol. 13, 889811. doi:10.3389/fmicb.2022.889811
Bar-On, Y. M., Flamholz, A., Phillips, R., and Milo, R. (2020). SARS-CoV-2 (COVID-19) by the numbers. Elife 9, e57309. doi:10.7554/elife.57309
Barreiro, A., Prenafeta, A., Bech-Sabat, G., Roca, M., Perozo Mur, E., March, R., et al. (2022). Preclinical efficacy, safety, and immunogenicity of a COVID-19 vaccine candidate based on a recombinant RBD fusion heterodimer of SARS-CoV-2. bioRxiv
Barrett, P. N., Mundt, W., Kistner, O., and Howard, M. K. (2009). Vero cell platform in vaccine production: Moving towards cell culture-based viral vaccines. Expert Rev. Vaccines 8, 607–618. doi:10.1586/erv.09.19
Bartik, A. W., Bertrand, M., Cullen, Z., Glaeser, E. L., Luca, M., and Stanton, C. (2020). The impact of COVID-19 on small business outcomes and expectations. Proc. Natl. Acad. Sci. U. S. A. 117, 17656–17666. doi:10.1073/pnas.2006991117
Barton, L. M., Duval, E. J., Stroberg, E., Ghosh, S., and Mukhopadhyay, S. (2020). COVID-19 autopsies, Oklahoma, USA. Am. J. Clin. Pathol. 153, 725–733. doi:10.1093/ajcp/aqaa062
Bartoszko, J. J., Farooqi, M. a. M., Alhazzani, W., and Loeb, M. (2020). Medical masks vs N95 respirators for preventing COVID-19 in healthcare workers: A systematic review and meta-analysis of randomized trials. Influenza Other Respir. Viruses 14, 365–373. doi:10.1111/irv.12745
Bartsch, S. M., O'shea, K. J., Chin, K. L., Strych, U., Ferguson, M. C., Bottazzi, M. E., et al. (2022). Maintaining face mask use before and after achieving different COVID-19 vaccination coverage levels: A modelling study. Lancet Public Health 7, e356–e365. doi:10.1016/s2468-2667(22)00040-8
Barycka, K., Szarpak, L., Filipiak, K. J., Jaguszewski, M., Smereka, J., Ladny, J. R., et al. (2020). Comparative effectiveness of N95 respirators and surgical/face masks in preventing airborne infections in the era of SARS-CoV2 pandemic: A meta-analysis of randomized trials. Plos One 15, e0242901. doi:10.1371/journal.pone.0242901
Bastard, P., Rosen, L. B., Zhang, Q., Michailidis, E., Hoffmann, H. H., Zhang, Y., et al. (2020). Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370, eabd4585. doi:10.1126/science.abd4585
Bastard, P., Gervais, A., Le Voyer, T., Rosain, J., Philippot, Q., Manry, J., et al. (2021). Autoantibodies neutralizing type I IFNs are present in ∼4% of uninfected individuals over 70 years old and account for ∼20% of COVID-19 deaths. Sci. Immunol. 6, eabl4340. doi:10.1126/sciimmunol.abl4340
Bauchner, H., Fontanarosa, P. B., and Livingston, E. H. (2020). Conserving supply of personal protective equipment—a call for ideas. JAMA 323, 1911. doi:10.1001/jama.2020.4770
Baz Morelli, A., Becher, D., Koernig, S., Silva, A., Drane, D., and Maraskovsky, E. (2012). Iscomatrix: A novel adjuvant for use in prophylactic and therapeutic vaccines against infectious diseases. J. Med. Microbiol. 61, 935–943. doi:10.1099/jmm.0.040857-0
Becerra-Flores, M., and Cardozo, T. (2020). SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int. J. Clin. Pract. 74, e13525. doi:10.1111/ijcp.13525
Beech, N., and Anseel, F. (2020). COVID-19 and its impact on management research and education: Threats, opportunities and a manifesto. Brit. J. Manage. 31, 447–449. doi:10.1111/1467-8551.12421
Beesoon, S., Behary, N., and Perwuelz, A. (2020). Universal masking during COVID-19 pandemic: Can textile engineering help public health? Narrative review of the evidence. Prev. Med. 139, 106236. doi:10.1016/j.ypmed.2020.106236
Bengtsson, K. L., Karlsson, K. H., Magnusson, S. E., Reimer, J. M., and Stertman, L. (2013). Matrix-M adjuvant: Enhancing immune responses by 'setting the stage' for the antigen. Expert Rev. Vaccines 12, 821–823. doi:10.1586/14760584.2013.814822
Bergamaschi, L., Mescia, F., Turner, L., Hanson, A. L., Kotagiri, P., Dunmore, B. J., et al. (2021). Longitudinal analysis reveals that delayed bystander CD8+ T cell activation and early immune pathology distinguish severe COVID-19 from mild disease. Immunity 54, 1257–1275. doi:10.1016/j.immuni.2021.05.010
Bergwerk, M., Gonen, T., Lustig, Y., Amit, S., Lipsitch, M., Cohen, C., et al. (2021). Covid-19 breakthrough infections in vaccinated health care workers. N. Engl. J. Med. Overseas. Ed. 385, 1474–1484. doi:10.1056/nejmoa2109072
Bestle, D., Heindl, M. R., Limburg, H., Van Lam Van, T., Pilgram, O., Moulton, H., et al. (2020). TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 3, e202000786. doi:10.26508/lsa.202000786
Bian, L., Gao, F., Zhang, J., He, Q., Mao, Q., Xu, M., et al. (2021). Effects of SARS-CoV-2 variants on vaccine efficacy and response strategies. Expert Rev. Vaccines 20, 365–373. doi:10.1080/14760584.2021.1903879
Binder, R. A., Alarja, N. A., Robie, E. R., Kochek, K. E., Xiu, L., Rocha-Melogno, L., et al. (2020). Environmental and aerosolized severe acute respiratory syndrome coronavirus 2 among hospitalized coronavirus disease 2019 patients. J. Infect. Dis. 222, 1798–1806. doi:10.1093/infdis/jiaa575
Birkhead, M., Glass, A. J., Allan-Gould, H., Goossens, C., and Wright, C. A. (2021). Ultrastructural evidence for vertical transmission of SARS-CoV-2. Int. J. Infect. Dis. 111, 10–11. doi:10.1016/j.ijid.2021.08.020
Biswas, C., Zhang, Y., Decastro, R., Guo, H., Nakamura, T., Kataoka, H., et al. (1995). The human tumor cell-derived collagenase stimulatory factor (renamed EMMPRIN) is a member of the immunoglobulin superfamily. Cancer Res. 55, 434–439.
Bizzarro, M. J., Conrad, S. A., Kaufman, D. A., and Rycus, P. (2011). Infections acquired during extracorporeal membrane oxygenation in neonates, children, and adults. Pediatr. Crit. Care Med. 12, 277–281. doi:10.1097/pcc.0b013e3181e28894
Block, S. L., Nolan, T., Sattler, C., Barr, E., Giacoletti, K. E., Marchant, C. D., et al. (2006). Comparison of the immunogenicity and reactogenicity of a prophylactic quadrivalent human papillomavirus (types 6, 11, 16, and 18) L1 virus-like particle vaccine in male and female adolescents and young adult women. Pediatrics 118, 2135–2145. doi:10.1542/peds.2006-0461
Boehm, E., Kronig, I., Neher, R. A., Eckerle, I., Vetter, P., and Kaiser, L. (2021). Novel SARS-CoV-2 variants: The pandemics within the pandemic. Clin. Microbiol. Infect. 27, 1109–1117. doi:10.1016/j.cmi.2021.05.022
Bollavaram, K., Leeman, T. H., Lee, M. W., Kulkarni, A., Upshaw, S. G., Yang, J., et al. (2021). Multiple sites on SARS-CoV-2 spike protein are susceptible to proteolysis by cathepsins B, K, L, S, and V. Protein Sci. 30, 1131–1143. doi:10.1002/pro.4073
Bonato, G., Dioscoridi, L., and Mutignani, M. (2020). Fecal-oral transmission of SARS-COV-2: Practical implications. Gastroenterology 159, 1621–1622. doi:10.1053/j.gastro.2020.03.066
Bos, R., Rutten, L., Van Der Lubbe, J. E. M., Bakkers, M. J. G., Hardenberg, G., Wegmann, F., et al. (2020). Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 Spike immunogen induces potent humoral and cellular immune responses. NPJ Vaccines 5, 91. doi:10.1038/s41541-020-00243-x
Bougakov, D., Podell, K., and Goldberg, E. (2021). Multiple neuroinvasive pathways in COVID-19. Mol. Neurobiol. 58, 564–575. doi:10.1007/s12035-020-02152-5
Bourouiba, L. (2020). Turbulent gas clouds and respiratory pathogen emissions: Potential implications for reducing transmission of COVID-19. JAMA 323, 1837–1838. doi:10.1001/jama.2020.4756
Brainard, J., Jones, N. R., Lake, I. R., Hooper, L., and Hunter, P. R. (2020). Community use of face masks and similar barriers to prevent respiratory illness such as COVID-19: A rapid scoping review. Eurosurveillance 25 (49), 2000725. doi:10.2807/1560-7917.ES.2020.25.49.2000725
Brammer, S., and Clark, T. (2020). COVID-19 and management education: Reflections on challenges, opportunities, and potential futures. Brit. J. Manage. 31, 453–456. doi:10.1111/1467-8551.12425
Brann, D. H., Tsukahara, T., Weinreb, C., Lipovsek, M., Van Den Berge, K., Gong, B., et al. (2020). Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 6, eabc5801. doi:10.1126/sciadv.abc5801
Brejová, B., Boršová, K., Hodorová, V., Čabanová, V., Gafurov, A., Fričová, D., et al. (2021). Nanopore sequencing of SARS-CoV-2: Comparison of short and long PCR-tiling amplicon protocols. Plos One 16, e0259277. doi:10.1371/journal.pone.0259277
Brouwer, P. J. M., Caniels, T. G., Van Der Straten, K., Snitselaar, J. L., Aldon, Y., Bangaru, S., et al. (2020). Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369, 643–650. doi:10.1126/science.abc5902
Brüssow, H., and Zuber, S. (2022). Can a combination of vaccination and face mask wearing contain the COVID-19 pandemic? Microb. Biotechnol. 15, 721–737. doi:10.1111/1751-7915.13997
Buckeridge, D., and Cadieux, G. (2006). Surveillance for newly emerging viruses. Perspect. Med. Virol. 16, 325–343. doi:10.1016/S0168-7069(06)16013-9
Bueckert, M., Gupta, R., Gupta, A., Garg, M., and Mazumder, A. (2020). Infectivity of SARS-CoV-2 and other coronaviruses on dry surfaces: Potential for indirect transmission. Mater. (Basel) 13, 5211. doi:10.3390/ma13225211
Bulbake, U., Doppalapudi, S., Kommineni, N., and Khan, W. (2017). Liposomal formulations in clinical use: An updated review. Pharmaceutics 9, 12. doi:10.3390/pharmaceutics9020012
Bull, R. A., Adikari, T. N., Ferguson, J. M., Hammond, J. M., Stevanovski, I., Beukers, A. G., et al. (2020). Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat. Commun. 11, 6272. doi:10.1038/s41467-020-20075-6
Cai, L., Liu, C., Fan, G., Liu, C., and Sun, X. (2019). Preventing viral disease by ZnONPs through directly deactivating TMV and activating plant immunity in Nicotiana benthamiana. Environ. Sci. Nano 6, 3653–3669. doi:10.1039/c9en00850k
Caldas, L. A., Carneiro, F. A., Higa, L. M., Monteiro, F. L., Da Silva, G. P., Da Costa, L. J., et al. (2020). Ultrastructural analysis of SARS-CoV-2 interactions with the host cell via high resolution scanning electron microscopy. Sci. Rep. 10, 16099. doi:10.1038/s41598-020-73162-5
Callaway, E. (2020). The race for coronavirus vaccines: A graphical guide. Nature 580, 576–577. doi:10.1038/d41586-020-01221-y
Camp, J. V., Buchta, C., Jovanovic, J., Puchhammer-Stockl, E., Benka, B., Griesmacher, A., et al. (2021). RT-PCR based SARS-CoV-2 variant screening assays require careful quality control. J. Clin. Virol. 141, 104905. doi:10.1016/j.jcv.2021.104905
Campos, E. V. R., Pereira, A. E. S., De Oliveira, J. L., Carvalho, L. B., Guilger-Casagrande, M., De Lima, R., et al. (2020). How can nanotechnology help to combat COVID-19? Opportunities and urgent need. J. Nanobiotechnology 18, 125. doi:10.1186/s12951-020-00685-4
Caniels, T. G., Bontjer, I., Van Der Straten, K., Poniman, M., Burger, J. A., Appelman, B., et al. (2021). Emerging SARS-CoV-2 variants of concern evade humoral immune responses from infection and vaccination. Sci. Adv. 7, eabj5365. doi:10.1126/sciadv.abj5365
Cantuti-Castelvetri, L., Ojha, R., Pedro, L. D., Djannatian, M., Franz, J., Kuivanen, S., et al. (2020). Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370, 856–860. doi:10.1126/science.abd2985
Cao, X., Ye, Y., and Liu, S. (2011). Gold nanoparticle-based signal amplification for biosensing. Anal. Biochem. 417, 1–16. doi:10.1016/j.ab.2011.05.027
Cao, Y., Su, B., Guo, X., Sun, W., Deng, Y., Bao, L., et al. (2020). Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell 182, 73–84.e16. e16. doi:10.1016/j.cell.2020.05.025
Cao, Y., Wang, J., Jian, F., Xiao, T., Song, W., Yisimayi, A., et al. (2022). Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602, 657–663. doi:10.1038/s41586-021-04385-3
Carraturo, F., Del Giudice, C., Morelli, M., Cerullo, V., Libralato, G., Galdiero, E., et al. (2020). Persistence of SARS-CoV-2 in the environment and COVID-19 transmission risk from environmental matrices and surfaces. Environ. Pollut. 265, 115010. doi:10.1016/j.envpol.2020.115010
Carvalho, A., Alqusairi, R., Adams, A., Paul, M., Kothari, N., Peters, S., et al. (2020). SARS-CoV-2 gastrointestinal infection causing hemorrhagic colitis: Implications for detection and transmission of COVID-19 disease. Am. J. Gastroenterol. 115, 942–946. doi:10.14309/ajg.0000000000000667
Castano-Jaramillo, L. M., Yamazaki-Nakashimada, M. A., O'farrill-Romanillos, P. M., Muzquiz Zermeño, D., Scheffler Mendoza, S. C., Venegas Montoya, E., et al. (2021). COVID-19 in the context of inborn errors of immunity: A case series of 31 patients from Mexico. J. Clin. Immunol. 41, 1463–1478. doi:10.1007/s10875-021-01077-5
Catching, A., Capponi, S., Yeh, M. T., Bianco, S., and Andino, R. (2021). Examining the interplay between face mask usage, asymptomatic transmission, and social distancing on the spread of COVID-19. Sci. Rep. 11, 15998. doi:10.1038/s41598-021-94960-5
Cavalcanti, I. D. L., and Cajubá De Britto Lira Nogueira, M. (2020). Pharmaceutical nanotechnology: Which products are been designed against COVID-19? J. Nanopart. Res. 22, 276. doi:10.1007/s11051-020-05010-6
Cavanagh, D. (1995). “The coronavirus surface glycoprotein,” in The coronaviridae. Editor S. G. Siddell (Boston, MA: Springer US), 73–113.
Cevik, M., Tate, M., Lloyd, O., Maraolo, A. E., Schafers, J., and Ho, A. (2021). SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: A systematic review and meta-analysis. Lancet Microbe 2, e13–e22. doi:10.1016/s2666-5247(20)30172-5
Chacón-Torres, J. C., Reinoso, C., Navas-León, D. G., Briceño, S., and González, G. (2020). Optimized and scalable synthesis of magnetic nanoparticles for RNA extraction in response to developing countries' needs in the detection and control of SARS-CoV-2. Sci. Rep. 10, 19004. doi:10.1038/s41598-020-75798-9
Chahal, J. S., Khan, O. F., Cooper, C. L., Mcpartlan, J. S., Tsosie, J. K., Tilley, L. D., et al. (2016). Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Natl. Acad. Sci. U. S. A. 113, E4133–E4142. doi:10.1073/pnas.1600299113
Chan, J. F., Kok, K. H., Zhu, Z., Chu, H., To, K. K., Yuan, S., et al. (2020). Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 9, 221–236. doi:10.1080/22221751.2020.1719902
Chan, T., Klaus, J., Meli, M. L., and Hofmann-Lehmann, R. (2021). SARS-CoV-2 infections in cats, dogs, and other animal species: Findings on infection and data from Switzerland. Schweiz Arch. Tierheilkd 163, 821–835. doi:10.17236/sat00329
Chan, J. F., Siu, G. K., Yuan, S., Ip, J. D., Cai, J. P., Chu, A. W., et al. (2022). Probable animal-to-human transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Delta variant AY.127 causing a pet shop-related coronavirus disease 2019 (COVID-19) outbreak in Hong Kong. Clin. Infect. Dis. 75, e76–e81. doi:10.1093/cid/ciac171
Chang, L., Zhao, L., Gong, H., Wang, L., and Wang, L. (2020). Severe acute respiratory syndrome coronavirus 2 RNA detected in blood donations. Emerg. Infect. Dis. 26, 1631–1633. doi:10.3201/eid2607.200839
Chang, S. Y., Huang, K. Y., Chao, T. L., Kao, H. C., Pang, Y. H., Lu, L., et al. (2021). Nanoparticle composite TPNT1 is effective against SARS-CoV-2 and influenza viruses. Sci. Rep. 11, 8692. doi:10.1038/s41598-021-87254-3
Chang, D., Chang, X., He, Y., and Tan, K. J. K. (2022). The determinants of COVID-19 morbidity and mortality across countries. Sci. Rep. 12, 5888. doi:10.1038/s41598-022-09783-9
Chau, N. V. V., Hong, N. T. T., Ngoc, N. M., Thanh, T. T., Khanh, P. N. Q., Nguyet, L. A., et al. (2021). Superspreading event of SARS-CoV-2 infection at a bar, Ho chi minh city, vietnam. Emerg. Infect. Dis. 27, 310–314. doi:10.3201/eid2701.203480
Chaubey, I., Vignesh, R., Babu, H., Wagoner, I., Govindaraj, S., and Velu, V. (2021). SARS-CoV-2 in pregnant women: Consequences of vertical transmission. Front. Cell. Infect. Microbiol. 11, 717104. doi:10.3389/fcimb.2021.717104
Chaudhary, V., Royal, A., Chavali, M., and Yadav, S. K. (2021). Advancements in research and development to combat COVID-19 using nanotechnology. Nanotechnol. Environ. Eng. 6, 8. doi:10.1007/s41204-021-00102-7
Chauhan, G., Madou, M. J., Kalra, S., Chopra, V., Ghosh, D., and Martinez-Chapa, S. O. (2020). Nanotechnology for COVID-19: Therapeutics and vaccine research. ACS Nano 14, 7760–7782. doi:10.1021/acsnano.0c04006
Chen, M., Shen, W., Rowan, N. R., Kulaga, H., Hillel, A., Ramanathan, M., et al. (2020). Elevated ACE-2 expression in the olfactory neuroepithelium: Implications for anosmia and upper respiratory SARS-CoV-2 entry and replication. Eur. Respir. J. 56, 2001948. doi:10.1183/13993003.01948-2020
Chen, J., Wang, R., Gilby, N. B., and Wei, G. W. (2021a). Omicron (B.1.1.529): Infectivity, vaccine breakthrough, and antibody resistance. ArXiv 62 (2), 412–422. arXiv:2112.01318v1.
Chen, M., Yuan, Y., Zhou, Y., Deng, Z., Zhao, J., Feng, F., et al. (2021b). Safety of SARS-CoV-2 vaccines: A systematic review and meta-analysis of randomized controlled trials. Infect. Dis. Poverty 10, 94. doi:10.1186/s40249-021-00878-5
Chen, W. H., Wei, J., Kundu, R. T., Adhikari, R., Liu, Z., Lee, J., et al. (2021c). Genetic modification to design a stable yeast-expressed recombinant SARS-CoV-2 receptor binding domain as a COVID-19 vaccine candidate. Biochimica Biophysica Acta - General Subj. 1865, 129893. doi:10.1016/j.bbagen.2021.129893
Cheng, M. P., Papenburg, J., Desjardins, M., Kanjilal, S., Quach, C., Libman, M., et al. (2020a). Diagnostic testing for severe acute respiratory syndrome-related coronavirus 2: A narrative review. Ann. Intern. Med. 172, 726–734. doi:10.7326/m20-1301
Cheng, Y., Luo, R., Wang, K., Zhang, M., Wang, Z., Dong, L., et al. (2020b). Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 97, 829–838. doi:10.1016/j.kint.2020.03.005
Chenthamara, D., Subramaniam, S., Ramakrishnan, S. G., Krishnaswamy, S., Essa, M. M., Lin, F. H., et al. (2019). Therapeutic efficacy of nanoparticles and routes of administration. Biomater. Res. 23, 20. doi:10.1186/s40824-019-0166-x
Cheung, K. S., Hung, I. F. N., Chan, P. P. Y., Lung, K. C., Tso, E., Liu, R., et al. (2020). Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: Systematic review and meta-analysis. Gastroenterology 159, 81–95. doi:10.1053/j.gastro.2020.03.065
Chiale, C., Greene, T. T., and Zuniga, E. I. (2022). Interferon induction, evasion, and paradoxical roles during SARS-CoV-2 infection. Immunol. Rev. 309, 12–24. doi:10.1111/imr.13113
China-National-Health-Commission-(Beijing) (2020). Announcement from the Headquarter for novel coronavirus pneumonia prevention and control.
Chu, D. K., Akl, E. A., Duda, S., Solo, K., Yaacoub, S., Schünemann, H. J., et al. (2020). Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: A systematic review and meta-analysis. Lancet 395, 1973–1987. doi:10.1016/s0140-6736(20)31142-9
Chung, Y. H., Beiss, V., Fiering, S. N., and Steinmetz, N. F. (2020). COVID-19 vaccine frontrunners and their nanotechnology design. ACS Nano 14, 12522–12537. doi:10.1021/acsnano.0c07197
Cibulski, S. P., Mourglia-Ettlin, G., Teixeira, T. F., Quirici, L., Roehe, P. M., Ferreira, F., et al. (2016). Novel ISCOMs from Quillaja brasiliensis saponins induce mucosal and systemic antibody production, T-cell responses and improved antigen uptake. Vaccine 34, 1162–1171. doi:10.1016/j.vaccine.2016.01.029
Ciuffreda, L., Lorenzo-Salazar, J. M., Alcoba-Florez, J., Rodriguez-Perez, H., Gil-Campesino, H., Inigo-Campos, A., et al. (2021a). Longitudinal study of a SARS-CoV-2 infection in an immunocompromised patient with X-linked agammaglobulinemia. J. Infect. 83, 607–635. doi:10.1016/j.jinf.2021.07.028
Ciuffreda, L., Rodriguez-Perez, H., and Flores, C. (2021b). Nanopore sequencing and its application to the study of microbial communities. Comput. Struct. Biotechnol. J. 19, 1497–1511. doi:10.1016/j.csbj.2021.02.020
Ciuffreda, L., Alcoba-Florez, J., Lorenzo-Salazar, J. M., Gil-Campesino, H., Garcia-Martinez De Artola, D., Diez-Gil, O., et al. (2022). Association of the Delta SARS-CoV-2 variant with 28-day hospital mortality between december 2020 and september 2021. J. Infect. 85, 90–122. doi:10.1016/j.jinf.2022.04.030
Cleary, S. M., Wilkinson, T., Tamandjou Tchuem, C. R., Docrat, S., and Solanki, G. C. (2021). Cost-effectiveness of intensive care for hospitalized COVID-19 patients: Experience from south Africa. BMC Health Serv. Res. 21, 82. doi:10.1186/s12913-021-06081-4
Clinkard, D., Mashari, A., Karkouti, K., and Fedorko, L. (2021). Evaluation of N95 respirators, modified snorkel masks and low-cost powered air-purifying respirators: A prospective observational cohort study in healthcare workers. Anaesthesia 76, 617–622. doi:10.1111/anae.15392
Coloma, J., Jain, R., Rajashankar, K. R., García-Sastre, A., and Aggarwal, A. K. (2016). Structures of NS5 methyltransferase from Zika virus. Cell Rep. 16, 3097–3102. doi:10.1016/j.celrep.2016.08.091
Comber, J. D., and Philip, R. (2014). MHC class I antigen presentation and implications for developing a new generation of therapeutic vaccines. Ther. Adv. Vaccines 2, 77–89. doi:10.1177/2051013614525375
Constantin, C., Pisani, A., Bardi, G., and Neagu, M. (2021). Nano-carriers of COVID-19 vaccines: The main pillars of efficacy. Nanomedicine (Lond) 16, 2377–2387. doi:10.2217/nnm-2021-0250
Conti, M. G., Terreri, S., Piano Mortari, E., Albano, C., Natale, F., Boscarino, G., et al. (2021). Immune response of neonates born to mothers infected with SARS-CoV-2. JAMA Netw. Open 4, e2132563. doi:10.1001/jamanetworkopen.2021.32563
Corbett, K. S., Edwards, D. K., Leist, S. R., Abiona, O. M., Boyoglu-Barnum, S., Gillespie, R. A., et al. (2020). SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571. doi:10.1038/s41586-020-2622-0
Corbett, K. S., Nason, M. C., Flach, B., Gagne, M., O'connell, S., Johnston, T. S., et al. (2021). Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science 373, eabj0299. doi:10.1126/science.abj0299
Corman, V. M., Landt, O., Kaiser, M., Molenkamp, R., Meijer, A., Chu, D. K., et al. (2020). Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 25. doi:10.2807/1560-7917.es.2020.25.3.2000045
Corominas, J., Garriga, C., Prenafeta, A., Moros, A., Cañete, M., Barreiro, A., et al. (2022). Safety and immunogenicity of the protein-based PHH-1V compared to BNT162b2 as a heterologous SARS-CoV-2 booster vaccine in adults vaccinated against COVID-19: A multicentre, randomised, double-blind, non-inferiority phase IIb trial. medRxiv. doi:10.1101/2022.07.05.22277210
Costa, S., Giordano, L., Bottoni, A., Tiberi, E., Fattore, S., Pastorino, R., et al. (2022). Vertical transmission of SARS-CoV-2 during pregnancy: A prospective Italian cohort study. Am. J. Perinatol. doi:10.1055/a-1792-4535
Covid-19-Host-Genetics-Initiative (2021). The mutation that helps Delta spread like wildfire. Nature 600, 472–473. doi:10.1038/d41586-021-02275-2
Creech, C. B., Walker, S. C., and Samuels, R. J. (2021). SARS-CoV-2 vaccines. Jama 325, 1318–1320. doi:10.1001/jama.2021.3199
Cucinotta, D., and Vanelli, M. (2020). WHO declares COVID-19 a pandemic. Acta Biomed. 91, 157–160. doi:10.23750/abm.v91i1.9397
Cuicchi, D., Lazzarotto, T., and Poggioli, G. (2021). Fecal-oral transmission of SARS-CoV-2: Review of laboratory-confirmed virus in gastrointestinal system. Int. J. Colorectal Dis. 36, 437–444. doi:10.1007/s00384-020-03785-7
Cyster, J. G., and Allen, C. D. C. (2019). B cell responses: Cell interaction dynamics and decisions. Cell 177, 524–540. doi:10.1016/j.cell.2019.03.016
Dai, L., and Gao, G. F. (2021). Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 21, 73–82. doi:10.1038/s41577-020-00480-0
Dai, L., Zheng, T., Xu, K., Han, Y., Xu, L., Huang, E., et al. (2020). A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell 182, 722–733.e11. e711. doi:10.1016/j.cell.2020.06.035
Dalvie, N. C., Rodriguez-Aponte, S. A., Hartwell, B. L., Tostanoski, L. H., Biedermann, A. M., Crowell, L. E., et al. (2021a). Engineered SARS-CoV-2 receptor binding domain improves manufacturability in yeast and immunogenicity in mice. Proc. Natl. Acad. Sci. U. S. A. 118, e2106845118. doi:10.1073/pnas.2106845118
Dalvie, N. C., Tostanoski, L. H., Rodriguez-Aponte, S. A., Kaur, K., Bajoria, S., Kumru, O. S., et al. (2021b). A modular protein subunit vaccine candidate produced in yeast confers protection against SARS-CoV-2 in non-human primates. bioRxiv., 452251. doi:10.1101/2021.07.13.452251
Daly, J. L., Simonetti, B., Klein, K., Chen, K. E., Williamson, M. K., Antón-Plágaro, C., et al. (2020). Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370, 861–865. doi:10.1126/science.abd3072
Daniloski, Z., Jordan, T. X., Ilmain, J. K., Guo, X., Bhabha, G., Tenoever, B. R., et al. (2021). The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. eLife 10, e65365. doi:10.7554/elife.65365
Day, M. (2020). Covid-19: Four fifths of cases are asymptomatic, China figures indicate. BMJ 369, m1375. doi:10.1136/bmj.m1375
De Alwis, R., Gan, E. S., Chen, S., Leong, Y. S., Tan, H. C., Zhang, S. L., et al. (2021). A single dose of self-transcribing and replicating RNA-based SARS-CoV-2 vaccine produces protective adaptive immunity in mice. Mol. Ther. 29, 1970–1983. doi:10.1016/j.ymthe.2021.04.001
De Groot, R. J., Luytjes, W., Horzinek, M. C., Van Der Zeijst, B. A., Spaan, W. J., and Lenstra, J. A. (1987). Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J. Mol. Biol. 196, 963–966. doi:10.1016/0022-2836(87)90422-0
De Nardo, P., Gentilotti, E., Mazzaferri, F., Cremonini, E., Hansen, P., Goossens, H., et al. (2020). Multi-Criteria Decision Analysis to prioritize hospital admission of patients affected by COVID-19 in low-resource settings with hospital-bed shortage. Int. J. Infect. Dis. 98, 494–500. doi:10.1016/j.ijid.2020.06.082
De Oliveira-Filho, E. F., De Carvalho, O. V., Carneiro, I. O., Fernandes, F. D., Vaz, S. N., Pedroso, C., et al. (2022). Frequent infection of cats with SARS-CoV-2 irrespective of pre-existing enzootic coronavirus immunity, Brazil 2020. Front. Immunol. 13, 857322. doi:10.3389/fimmu.2022.857322
Deamer, D., Akeson, M., and Branton, D. (2016). Three decades of nanopore sequencing. Nat. Biotechnol. 34, 518–524. doi:10.1038/nbt.3423
Debnath, S. K., and Srivastava, R. (2022). Potential application of bionanoparticles to treat severe acute respiratory syndrome coronavirus-2 infection. Front. Nanotechnol. 3. doi:10.3389/fnano.2021.813847
Della Cioppa, G., Nicolay, U., Lindert, K., Leroux-Roels, G., Clement, F., Castellino, F., et al. (2012). Superior immunogenicity of seasonal influenza vaccines containing full dose of MF59 ((R)) adjuvant: Results from a dose-finding clinical trial in older adults. Hum. Vaccin. Immunother. 8, 216–227. doi:10.4161/hv.18445
Della Cioppa, G., Nicolay, U., Lindert, K., Leroux-Roels, G., Clement, F., Castellino, F., et al. (2014). A dose-ranging study in older adults to compare the safety and immunogenicity profiles of MF59®-adjuvanted and non-adjuvanted seasonal influenza vaccines following intradermal and intramuscular administration. Hum. Vaccin. Immunother. 10, 1701–1710. doi:10.4161/hv.28618
Desai, A. D., Lavelle, M., Boursiquot, B. C., and Wan, E. Y. (2022). Long-term complications of COVID-19. Am. J. Physiology-Cell Physiology 322, C1–c11. doi:10.1152/ajpcell.00375.2021
Devaux, C. A., Rolain, J. M., and Raoult, D. (2020). ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 53, 425–435. doi:10.1016/j.jmii.2020.04.015
Dhama, K., Patel, S. K., Kumar, R., Masand, R., Rana, J., Yatoo, M. I., et al. (2021). The role of disinfectants and sanitizers during COVID-19 pandemic: Advantages and deleterious effects on humans and the environment. Environ. Sci. Pollut. Res. 28, 34211–34228. doi:10.1007/s11356-021-14429-w
Dheda, K., Charalambous, S., Karat, A. S., Von Delft, A., Lalloo, U. G., Van Zyl Smit, R., et al. (2021). A position statement and practical guide to the use of particulate filtering facepiece respirators (N95, FFP2, or equivalent) for South African health workers exposed to respiratory pathogens including Mycobacterium tuberculosis and SARS-CoV-2. Afr. J. Thorac. Crit. Care Med. 27. doi:10.7196/ajtccm.2021.v27i4.173
Di Fusco, M., Shea, K. M., Lin, J., Nguyen, J. L., Angulo, F. J., Benigno, M., et al. (2021). Health outcomes and economic burden of hospitalized COVID-19 patients in the United States. J. Med. Econ. 24, 308–317. doi:10.1080/13696998.2021.1886109
Diao, B., Wang, C., Tan, Y., Chen, X., Liu, Y., Ning, L., et al. (2020). Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front. Immunol. 11, 827. doi:10.3389/fimmu.2020.00827
Ding, Z., Qian, H., Xu, B., Huang, Y., Miao, T., Yen, H. L., et al. (2021). Toilets dominate environmental detection of severe acute respiratory syndrome coronavirus 2 in a hospital. Sci. Total Environ. 753, 141710. doi:10.1016/j.scitotenv.2020.141710
Dinnes, J., Deeks, J. J., Adriano, A., Berhane, S., Davenport, C., Dittrich, S., et al. (2020). Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 8, Cd013705. doi:10.1002/14651858.CD013705
Dolgin, E. (2021). CureVac COVID vaccine let-down spotlights mRNA design challenges. Nature 594, 483. doi:10.1038/d41586-021-01661-0
Dona, D., Minotti, C., Costenaro, P., Da Dalt, L., and Giaquinto, C. (2020). Fecal-oral transmission of SARS-CoV-2 in children: Is it time to change our approach? Pediatr. Infect. Dis. J. 39, e133–e134. doi:10.1097/inf.0000000000002704
Donaldson, B., Lateef, Z., Walker, G. F., Young, S. L., and Ward, V. K. (2018). Virus-like particle vaccines: Immunology and formulation for clinical translation. Expert Rev. Vaccines 17, 833–849. doi:10.1080/14760584.2018.1516552
Donskyi, I. S., Nie, C., Ludwig, K., Trimpert, J., Ahmed, R., Quaas, E., et al. (2021). SARS-CoV-2 inhibitors: Graphene sheets with defined dual functionalities for the strong SARS-CoV-2 interactions (small 11/2021). Small 17, 2170046. doi:10.1002/smll.202170046
Drożdżal, S., Rosik, J., Lechowicz, K., Machaj, F., Szostak, B., Przybyciński, J., et al. (2021). An update on drugs with therapeutic potential for SARS-CoV-2 (COVID-19) treatment. Drug resist. updat. 59, 100794. doi:10.1016/j.drup.2021.100794
Du, L., Zhao, G., Lin, Y., Sui, H., Chan, C., Ma, S., et al. (2008). Intranasal vaccination of recombinant adeno-associated virus encoding receptor-binding domain of severe acute respiratory syndrome coronavirus (SARS-CoV) spike protein induces strong mucosal immune responses and provides long-term protection against SARS-CoV infection. J. Immunol. 180, 948–956. doi:10.4049/jimmunol.180.2.948
Dumitriu, D., Emeruwa, U. N., Hanft, E., Liao, G. V., Ludwig, E., Walzer, L., et al. (2021). Outcomes of neonates born to mothers with severe acute respiratory syndrome coronavirus 2 infection at a large medical center in New York city. JAMA Pediatr. 175, 157–167. doi:10.1001/jamapediatrics.2020.4298
Dunkle, L. M., Kotloff, K. L., Gay, C. L., Áñez, G., Adelglass, J. M., Barrat Hernández, A. Q., et al. (2022). Efficacy and safety of NVX-CoV2373 in adults in the United States and Mexico. N. Engl. J. Med. Overseas. Ed. 386, 531–543. doi:10.1056/nejmoa2116185
Dutta, D., Naiyer, S., Mansuri, S., Soni, N., Singh, V., Bhat, K. H., et al. (2022). COVID-19 diagnosis: A comprehensive review of the RT-qPCR method for detection of SARS-CoV-2. Diagn. (Basel) 12, 1503. doi:10.3390/diagnostics12061503
Eaaswarkhanth, M., Al Madhoun, A., and Al-Mulla, F. (2020). Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? Int. J. Infect. Dis. 96, 459–460. doi:10.1016/j.ijid.2020.05.071
Earle, K. A., Ambrosino, D. M., Fiore-Gartland, A., Goldblatt, D., Gilbert, P. B., Siber, G. R., et al. (2021). Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine 39, 4423–4428. doi:10.1016/j.vaccine.2021.05.063
Elbe, S., and Buckland-Merrett, G. (2017). Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob. Chall. 1, 33–46. doi:10.1002/gch2.1018
Ella, R., Reddy, S., Blackwelder, W., Potdar, V., Yadav, P., Sarangi, V., et al. (2021a). Efficacy, safety, and lot-to-lot immunogenicity of an inactivated SARS-CoV-2 vaccine (BBV152): Interim results of a randomised, double-blind, controlled, phase 3 trial. Lancet 398, 2173–2184. doi:10.1016/S0140-6736(21)02000-6
Ella, R., Reddy, S., Jogdand, H., Sarangi, V., Ganneru, B., Prasad, S., et al. (2021b). Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: Interim results from a double-blind, randomised, multicentre, phase 2 trial, and 3-month follow-up of a double-blind, randomised phase 1 trial. Lancet Infect. Dis. 21, 950–961. doi:10.1016/s1473-3099(21)00070-0
Ellinghaus, D., Degenhardt, F., Bujanda, L., Buti, M., Albillos, A., Invernizzi, P., et al. (2020). Genomewide association study of severe covid-19 with respiratory failure. N. Engl. J. Med. Overseas. Ed. 383, 1522–1534. doi:10.1056/nejmoa2020283
Erasmus, J. H., Khandhar, A. P., O'connor, M. A., Walls, A. C., Hemann, E. A., Murapa, P., et al. (2020). An Alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci. Transl. Med. 12, eabc9396. doi:10.1126/scitranslmed.abc9396
Eslami, H., and Jalili, M. (2020). The role of environmental factors to transmission of SARS-CoV-2 (COVID-19). Amb. Express 10, 92. doi:10.1186/s13568-020-01028-0
Esposito, S., Principi, N., Leung, C. C., and Migliori, G. B. (2020). Universal use of face masks for success against COVID-19: Evidence and implications for prevention policies. Eur. Respir. J. 55, 2001260. doi:10.1183/13993003.01260-2020
Eugenia Toledo-Romaní, M., García-Carmenate, M., Valenzuela-Silva, C., Baldoquín-Rodríguez, W., Martínez-Pérez, M., Rodríguez-González, M., et al. (2022). Safety and efficacy of the two doses conjugated protein-based SOBERANA-02 COVID-19 vaccine and of a heterologous three-dose combination with SOBERANA-PLUS: Double-blind, randomised, placebo-controlled phase 3 clinical trial. medRxiv. doi:10.1101/2021.10.31.21265703
Ewer, K. J., Barrett, J. R., Belij-Rammerstorfer, S., Sharpe, H., Makinson, R., Morter, R., et al. (2021). T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat. Med. 27, 270–278. doi:10.1038/s41591-020-01194-5
Ezoe, S., Palacpac, N. M. Q., Tetsutani, K., Yamamoto, K., Okada, K., Taira, M., et al. (2020). First-in-human randomised trial and follow-up study of Plasmodium falciparum blood-stage malaria vaccine BK-SE36 with CpG-ODN(K3). Vaccine 38, 7246–7257. doi:10.1016/j.vaccine.2020.09.056
Fakhruddin, K. S., Haiat, A., Ngo, H. C., Panduwawala, C., Chang, J. W. W., and Samaranayake, L. P. (2022). Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) viral positivity and their burden in saliva of asymptomatic carriers - a systematic review and meta-analysis. Acta Odontol. Scand. 80, 182–190. doi:10.1080/00016357.2021.1977385
Fallerini, C., Daga, S., Mantovani, S., Benetti, E., Picchiotti, N., Francisci, D., et al. (2021). Association of toll-like receptor 7 variants with life-threatening COVID-19 disease in males: Findings from a nested case-control study. Elife 10, e67569. doi:10.7554/eLife.67569
Falzone, L., Gattuso, G., Tsatsakis, A., Spandidos, D. A., and Libra, M. (2021). Current and innovative methods for the diagnosis of COVID-19 infection (Review). Int. J. Mol. Med. 47, 100. doi:10.3892/ijmm.2021.4933
Fauziah, N., Koesoemadinata, R. C., Andriyoko, B., Faridah, L., Riswari, S. F., Widyatmoko, L., et al. (2021). The performance of point-of-care antibody test for COVID-19 diagnosis in a tertiary hospital in Bandung, Indonesia. J. Infect. Dev. Ctries. 15, 237–241. doi:10.3855/jidc.13370
Fenizia, C., Galbiati, S., Vanetti, C., Vago, R., Clerici, M., Tacchetti, C., et al. (2021). SARS-CoV-2 entry: At the crossroads of CD147 and ACE2. Cells 10, 1434. doi:10.3390/cells10061434
Fenton, O. S., Olafson, K. N., Pillai, P. S., Mitchell, M. J., and Langer, R. (2018). Advances in biomaterials for drug delivery. Adv. Mat. 30, e1705328. doi:10.1002/adma.201705328
Ferretti, L., Wymant, C., Kendall, M., Zhao, L., Nurtay, A., Abeler-Dorner, L., et al. (2020). Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science 368, eabb6936. doi:10.1126/science.abb6936
Flemming, A. (2022). Omicron, the great escape artist. Nat. Rev. Immunol. 22, 75. doi:10.1038/s41577-022-00676-6
Focosi, D., Maggi, F., and Casadevall, A. (2022). Mucosal vaccines, sterilizing immunity, and the future of SARS-CoV-2 virulence. Viruses 14, 187. doi:10.3390/v14020187
Folegatti, P. M., Ewer, K. J., Aley, P. K., Angus, B., Becker, S., Belij-Rammerstorfer, S., et al. (2020). Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 396, 467–478. doi:10.1016/S0140-6736(20)31604-4
Fonseca-Santos, B., Gremião, M. P., and Chorilli, M. (2015). Nanotechnology-based drug delivery systems for the treatment of Alzheimer's disease. Int. J. Nanomedicine 10, 4981–5003. doi:10.2147/IJN.S87148
Formica, N., Mallory, R., Albert, G., Robinson, M., Plested, J. S., Cho, I., et al. (2021). Different dose regimens of a SARS-CoV-2 recombinant spike protein vaccine (NVX-CoV2373) in younger and older adults: A phase 2 randomized placebo-controlled trial. PLoS Med. 18, e1003769. doi:10.1371/journal.pmed.1003769
Foster, J. B., Choudhari, N., Perazzelli, J., Storm, J., Hofmann, T. J., Jain, P., et al. (2019). Purification of mRNA encoding chimeric antigen receptor is critical for generation of a robust T-cell response. Hum. Gene Ther. 30, 168–178. doi:10.1089/hum.2018.145
Fougeroux, C., Goksøyr, L., Idorn, M., Soroka, V., Myeni, S. K., Dagil, R., et al. (2021). Capsid-like particles decorated with the SARS-CoV-2 receptor-binding domain elicit strong virus neutralization activity. Nat. Commun. 12, 324. doi:10.1038/s41467-020-20251-8
Fraser, B. J., Beldar, S., Seitova, A., Hutchinson, A., Mannar, D., Li, Y., et al. (2022). Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation. Nat. Chem. Biol. 18, 963–971. doi:10.1038/s41589-022-01059-7
Furuya-Kanamori, L., and Yakob, L. (2018). Heterogeneous and dynamic prevalence of asymptomatic influenza virus infections. Emerg. Infect. Dis. 24, 951. doi:10.3201/eid2405.180075
Gallo, O., Locatello, L. G., Mazzoni, A., Novelli, L., and Annunziato, F. (2021). The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS-CoV-2 infection. Mucosal Immunol. 14, 305–316. doi:10.1038/s41385-020-00359-2
Gandhi, M., and Rutherford, G. W. (2020). Facial masking for covid-19 - potential for "variolation" as we await a vaccine. N. Engl. J. Med. Overseas. Ed. 383, e101. doi:10.1056/nejmp2026913
Gangapurwala, G., Vollrath, A., De San Luis, A., and Schubert, U. S. (2020). PLA/PLGA-Based drug delivery systems produced with supercritical CO(2)-A green future for particle formulation? Pharmaceutics 12, 1118. doi:10.3390/pharmaceutics12111118
Gao, Q., Bao, L., Mao, H., Wang, L., Xu, K., Yang, M., et al. (2020a). Development of an inactivated vaccine candidate for SARS-CoV-2. Science 369, 77–81. doi:10.1126/science.abc1932
Gao, Y., Shi, C., Chen, Y., Shi, P., Liu, J., Xiao, Y., et al. (2020b). A cluster of the Corona Virus Disease 2019 caused by incubation period transmission in Wuxi, China. J. Infect. 80, 666–670. doi:10.1016/j.jinf.2020.03.042
Garcea, R. L., and Gissmann, L. (2004). Virus-like particles as vaccines and vessels for the delivery of small molecules. Curr. Opin. Biotechnol. 15, 513–517. doi:10.1016/j.copbio.2004.10.002
Gardy, J., Loman, N. J., and Rambaut, A. (2015). Real-time digital pathogen surveillance - the time is now. Genome Biol. 16, 155. doi:10.1186/s13059-015-0726-x
Gavriatopoulou, M., Korompoki, E., Fotiou, D., Ntanasis-Stathopoulos, I., Psaltopoulou, T., Kastritis, E., et al. (2020). Organ-specific manifestations of COVID-19 infection. Clin. Exp. Med. 20, 493–506. doi:10.1007/s10238-020-00648-x
Geng, Q., Tai, W., Baxter, V. K., Shi, J., Wan, Y., Zhang, X., et al. (2021). Novel virus-like nanoparticle vaccine effectively protects animal model from SARS-CoV-2 infection. PLoS Pathog. 17, e1009897. doi:10.1371/journal.ppat.1009897
Gengler, I., Wang, J. C., Speth, M. M., and Sedaghat, A. R. (2020). Sinonasal pathophysiology of SARS-CoV-2 and COVID-19: A systematic review of the current evidence. Laryngoscope Investig. Otolaryngol. 5, 354–359. doi:10.1002/lio2.384
George, M. P., Maier, L. A., Kasperbauer, S., Eddy, J., Mayer, A. S., and Magin, C. M. (2020). How to leverage collaborations between the BME community and local hospitals to address critical personal protective equipment shortages during the COVID-19 pandemic. Ann. Biomed. Eng. 48, 2281–2284. doi:10.1007/s10439-020-02580-3
Gholami, M., Fawad, I., Shadan, S., Rowaiee, R., Ghanem, H., Hassan Khamis, A., et al. (2021). COVID-19 and healthcare workers: A systematic review and meta-analysis. Int. J. Infect. Dis. 104, 335–346. doi:10.1016/j.ijid.2021.01.013
Gibson, P. G., Qin, L., and Puah, S. H. (2020). COVID-19 acute respiratory distress syndrome (ARDS): Clinical features and differences from typical pre-COVID-19 ARDS. Med. J. Aust. 213, 54–56. e51. doi:10.5694/mja2.50674
Glenet, M., Lebreil, A. L., Heng, L., N'guyen, Y., Meyer, I., and Andreoletti, L. (2021). Asymptomatic COVID-19 adult outpatients identified as significant viable SARS-CoV-2 shedders. Sci. Rep. 11, 20615. doi:10.1038/s41598-021-00142-8
Global-Preparedness-Monitoring-Board-(Gpmb) (2019). A world at risk: Annual report on global preparedness for health emergencies. Available at: https://apps.who.int/gpmb/assets/annual_report/GPMB_annualreport_2019.pdf?utm_source=mandiner&utm_medium=link&utm_campaign=mandiner_202004.
Goepfert, P. A., Fu, B., Chabanon, A. L., Bonaparte, M. I., Davis, M. G., Essink, B. J., et al. (2021). Safety and immunogenicity of SARS-CoV-2 recombinant protein vaccine formulations in healthy adults: Interim results of a randomised, placebo-controlled, phase 1-2, dose-ranging study. Lancet Infect. Dis. 21, 1257–1270. doi:10.1016/s1473-3099(21)00147-x
Goharshadi, E. K., Goharshadi, K., and Moghayedi, M. (2022). The use of nanotechnology in the fight against viruses: A critical review. Coord. Chem. Rev. 464, 214559. doi:10.1016/j.ccr.2022.214559
Gohl, D. M., Garbe, J., Grady, P., Daniel, J., Watson, R. H. B., Auch, B., et al. (2020). A rapid, cost-effective tailed amplicon method for sequencing SARS-CoV-2. BMC Genomics 21, 863. doi:10.1186/s12864-020-07283-6
Gonzalez, A., Aboubakr, H. A., Brockgreitens, J., Hao, W., Wang, Y., Goyal, S. M., et al. (2021). Durable nanocomposite face masks with high particulate filtration and rapid inactivation of coronaviruses. Sci. Rep. 11, 24318. doi:10.1038/s41598-021-03771-1
González-Recio, O., Gutiérrez-Rivas, M., Peiró-Pastor, R., Aguilera-Sepúlveda, P., Cano-Gómez, C., Jiménez-Clavero, M., et al. (2021). Sequencing of SARS-CoV-2 genome using different nanopore chemistries. Appl. Microbiol. Biotechnol. 105, 3225–3234. doi:10.1007/s00253-021-11250-w
Goyal, P., Choi, J. J., Pinheiro, L. C., Schenck, E. J., Chen, R., Jabri, A., et al. (2020). Clinical characteristics of covid-19 in New York city. N. Engl. J. Med. Overseas. Ed. 382, 2372–2374. doi:10.1056/nejmc2010419
Grasselli, G., Greco, M., Zanella, A., Albano, G., Antonelli, M., Bellani, G., et al. (2020). Risk factors associated with mortality among patients with COVID-19 in intensive care units in lombardy, Italy. JAMA Intern. Med. 180, 1345–1355. doi:10.1001/jamainternmed.2020.3539
Grassia, R., Testa, S., Pan, A., and Conti, C. B. (2020). SARS-CoV-2 and gastrointestinal tract: The dark side of the pandemic. Dig. Liver Dis. 52, 700–701. doi:10.1016/j.dld.2020.04.028
Grau-Exposito, J., Sanchez-Gaona, N., Massana, N., Suppi, M., Astorga-Gamaza, A., Perea, D., et al. (2021). Peripheral and lung resident memory T cell responses against SARS-CoV-2. Nat. Commun. 12, 3010. doi:10.1038/s41467-021-23333-3
Guan, W. J., Ni, Z. Y., Hu, Y., Liang, W. H., Ou, C. Q., He, J. X., et al. (2020). Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. Overseas. Ed. 382, 1708–1720. doi:10.1056/nejmoa2002032
Guebre-Xabier, M., Patel, N., Tian, J. H., Zhou, B., Maciejewski, S., Lam, K., et al. (2020). NVX-CoV2373 vaccine protects cynomolgus macaque upper and lower airways against SARS-CoV-2 challenge. Vaccine 38, 7892–7896. doi:10.1016/j.vaccine.2020.10.064
Guerrini, G., Magrì, D., Gioria, S., Medaglini, D., and Calzolai, L. (2022). Characterization of nanoparticles-based vaccines for COVID-19. Nat. Nanotechnol. 17, 570–576. doi:10.1038/s41565-022-01129-w
Guo, J., Wang, Y., Niu, S., Li, H., Tian, Y., Yu, S., et al. (2020a). Highly sensitive fluorescence-linked immunosorbent assay for the determination of human IgG in serum using quantum dot nanobeads and magnetic Fe(3)O(4) nanospheres. ACS Omega 5, 23229–23236. doi:10.1021/acsomega.0c02987
Guo, L., Ren, L., Yang, S., Xiao, M., ChangYang, F., Dela Cruz, C. S., et al. (2020b). Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin. Infect. Dis. 71, 778–785. doi:10.1093/cid/ciaa310
Guo, Z. D., Wang, Z. Y., Zhang, S. F., Li, X., Li, L., Li, C., et al. (2020c). Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, wuhan, China, 2020. Emerg. Infect. Dis. 26, 1583–1591. doi:10.3201/eid2607.200885
Gupta, R., Sagar, P., Priyadarshi, N., Kaul, S., Sandhir, R., Rishi, V., et al. (2020). Nanotechnology-based approaches for the detection of SARS-CoV-2. Front. Nanotechnol. 2. doi:10.3389/fnano.2020.589832
Gupta, N., Augustine, S., Narayan, T., O'riordan, A., Das, A., Kumar, D., et al. (2021a). Point-of-Care PCR assays for COVID-19 detection. Biosens. (Basel) 11, 141. doi:10.3390/bios11050141
Gupta, V., Bhoyar, R. C., Jain, A., Srivastava, S., Upadhayay, R., Imran, M., et al. (2021b). Asymptomatic reinfection in 2 healthcare workers from India with genetically distinct severe acute respiratory syndrome coronavirus 2. Clin. Infect. Dis. 73, e2823–e2825. doi:10.1093/cid/ciaa1451
Hachmann, N. P., Miller, J., Collier, A. Y., Ventura, J. D., Yu, J., Rowe, M., et al. (2022). Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. Overseas. Ed. 387, 86–88. doi:10.1056/nejmc2206576
Hadas, Y., Sultana, N., Youssef, E., Sharkar, M. T. K., Kaur, K., Chepurko, E., et al. (2019). Optimizing modified mRNA in vitro synthesis protocol for heart gene therapy. Mol. Ther. - Methods & Clin. Dev. 14, 300–305. doi:10.1016/j.omtm.2019.07.006
Hadfield, J., Megill, C., Bell, S. M., Huddleston, J., Potter, B., Callender, C., et al. (2018). Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123. doi:10.1093/bioinformatics/bty407
Hagan, L. M., Mccormick, D. W., Lee, C., Sleweon, S., Nicolae, L., Dixon, T., et al. (2021). Outbreak of SARS-CoV-2 B.1.617.2 (delta) variant infections among incarcerated persons in a federal prison - Texas, july-august 2021. MMWR. Morb. Mortal. Wkly. Rep. 70, 1349–1354. doi:10.15585/mmwr.mm7038e3
Hamming, I., Timens, W., Bulthuis, M. L., Lely, A. T., Navis, G., and Van Goor, H. (2004). Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203, 631–637. doi:10.1002/path.1570
Hamza, R. Z., Gobouri, A. A., Al-Yasi, H. M., Al-Talhi, T. A., and El-Megharbel, S. M. (2021). A new sterilization strategy using TiO2 nanotubes for production of free radicals that eliminate viruses and application of a treatment strategy to combat infections caused by emerging SARS-CoV-2 during the COVID-19 pandemic. Coatings 11, 680. doi:10.3390/coatings11060680
Han, B., Song, Y., Li, C., Yang, W., Ma, Q., Jiang, Z., et al. (2021). Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine (CoronaVac) in healthy children and adolescents: A double-blind, randomised, controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 21, 1645–1653. doi:10.1016/s1473-3099(21)00319-4
Hao, X., Cheng, S., Wu, D., Wu, T., Lin, X., and Wang, C. (2020). Reconstruction of the full transmission dynamics of COVID-19 in Wuhan. Nature 584, 420–424. doi:10.1038/s41586-020-2554-8
Hariri, L., and Hardin, C. C. (2020). Covid-19, angiogenesis, and ARDS endotypes. N. Engl. J. Med. Overseas. Ed. 383, 182–183. doi:10.1056/nejme2018629
Harrigan, S. P., Wilton, J., Chong, M., Abdia, Y., Garcia, H. V., Rose, C., et al. (2022). Clinical severity of omicron SARS-CoV-2 variant relative to delta in British columbia, Canada: A retrospective analysis of whole genome sequenced cases. Clin. Infect. Dis. 30, ciac705. doi:10.1093/cid/ciac705
Hasanoglu, I., Korukluoglu, G., Asilturk, D., Cosgun, Y., Kalem, A. K., Altas, A. B., et al. (2021). Higher viral loads in asymptomatic COVID-19 patients might be the invisible part of the iceberg. Infection 49, 117–126. doi:10.1007/s15010-020-01548-8
Hasanzadeh, A., Alamdaran, M., Ahmadi, S., Nourizadeh, H., Bagherzadeh, M. A., Mofazzal Jahromi, M. A., et al. (2021). Nanotechnology against COVID-19: Immunization, diagnostic and therapeutic studies. J. Control. Release 336, 354–374. doi:10.1016/j.jconrel.2021.06.036
Hayn, M., Hirschenberger, M., Koepke, L., Nchioua, R., Straub, J. H., Klute, S., et al. (2021). Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities. Cell Rep. 35, 109126. doi:10.1016/j.celrep.2021.109126
He, X., Lau, E. H. Y., Wu, P., Deng, X., Wang, J., Hao, X., et al. (2020). Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 26, 672–675. doi:10.1038/s41591-020-0869-5
He, L., Lin, X., Wang, Y., Abraham, C., Sou, C., Ngo, T., et al. (2021). Single-component, self-assembling, protein nanoparticles presenting the receptor binding domain and stabilized spike as SARS-CoV-2 vaccine candidates. Sci. Adv. 7, eabf1591. doi:10.1126/sciadv.abf1591
Heath, P. T., Galiza, E. P., Baxter, D. N., Boffito, M., Browne, D., Burns, F., et al. (2021). Safety and efficacy of NVX-CoV2373 covid-19 vaccine. N. Engl. J. Med. Overseas. Ed. 385, 1172–1183. doi:10.1056/nejmoa2107659
Heida, R., Hinrichs, W. L., and Frijlink, H. W. (2022). Inhaled vaccine delivery in the combat against respiratory viruses: A 2021 overview of recent developments and implications for COVID-19. Expert Rev. Vaccines 21, 957–974. doi:10.1080/14760584.2021.1903878
Heinberg, L. J., and Steffen, K. (2021). Social isolation and loneliness during the COVID-19 pandemic: Impact on weight. Curr. Obes. Rep. 10, 365–370. doi:10.1007/s13679-021-00447-9
Heinz, F. X., and Stiasny, K. (2021). Distinguishing features of current COVID-19 vaccines: Knowns and unknowns of antigen presentation and modes of action. NPJ Vaccines 6, 104. doi:10.1038/s41541-021-00369-6
Helms, J., Kremer, S., Merdji, H., Clere-Jehl, R., Schenck, M., Kummerlen, C., et al. (2020). Neurologic features in severe SARS-CoV-2 infection. N. Engl. J. Med. Overseas. Ed. 382, 2268–2270. doi:10.1056/nejmc2008597
Hill, A., Wang, J., Levi, J., Heath, K., and Fortunak, J. (2020). Minimum costs to manufacture new treatments for COVID-19. J. Virus Erad. 6, 61–69. doi:10.1016/s2055-6640(20)30018-2
Hirotsu, Y., and Omata, M. (2021). Discovery of a SARS-CoV-2 variant from the P.1 lineage harboring K417T/E484K/N501Y mutations in Kofu, Japan. J. Infect. 82, 276–316. doi:10.1016/j.jinf.2021.03.013
Hoang, T. H., Vu, G. M., Tran, M. H., Tran, T. T. H., Le, Q. D., Tran, K. V., et al. (2022). Investigation of target sequencing of SARS-CoV-2 and immunogenic GWAS profiling in host cells of COVID-19 in Vietnam. BMC Infect. Dis. 22, 558. doi:10.1186/s12879-022-07415-1
Hodgson, S. H., Mansatta, K., Mallett, G., Harris, V., Emary, K. R. W., and Pollard, A. J. (2021). What defines an efficacious COVID-19 vaccine? A review of the challenges assessing the clinical efficacy of vaccines against SARS-CoV-2. Lancet Infect. Dis. 21, e26–e35. doi:10.1016/s1473-3099(20)30773-8
Hoehl, S., Rabenau, H., Berger, A., Kortenbusch, M., Cinatl, J., Bojkova, D., et al. (2020). Evidence of SARS-CoV-2 infection in returning travelers from wuhan, China. N. Engl. J. Med. Overseas. Ed. 382, 1278–1280. doi:10.1056/nejmc2001899
Hoffmann, M., Kleine-Weber, H., and Pohlmann, S. (2020a). A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 78, 779–784.e5. doi:10.1016/j.molcel.2020.04.022
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020b). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8. doi:10.1016/j.cell.2020.02.052
Hoffmann, M., Arora, P., Gross, R., Seidel, A., Hornich, B. F., Hahn, A. S., et al. (2021). SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 184, 2384–2393.e12. doi:10.1016/j.cell.2021.03.036
Hofmann, H., Pyrc, K., Van Der Hoek, L., Geier, M., Berkhout, B., and Pöhlmann, S. (2005). Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 102, 7988–7993. doi:10.1073/pnas.0409465102
Holzinger, M., Le Goff, A., and Cosnier, S. (2014). Nanomaterials for biosensing applications: A review. Front. Chem. 2, 63. doi:10.3389/fchem.2014.00063
Horejs, C. (2021). Artificially intelligent nanopore for rapid SARS-CoV-2 testing. Nat. Rev. Mat. 6, 650. doi:10.1038/s41578-021-00346-4
Hosseini, M., Behzadinasab, S., Benmamoun, Z., and Ducker, W. A. (2021). The viability of SARS-CoV-2 on solid surfaces. Curr. Opin. Colloid Interface Sci. 55, 101481. doi:10.1016/j.cocis.2021.101481
Hou, Y. J., Okuda, K., Edwards, C. E., Martinez, D. R., Asakura, T., Dinnon, K. H., et al. (2020). SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182, 429–446.e14. doi:10.1016/j.cell.2020.05.042
Howard, J., Huang, A., Li, Z., Tufekci, Z., Zdimal, V., Van Der Westhuizen, H. M., et al. (2021). An evidence review of face masks against COVID-19. Proc. Natl. Acad. Sci. U. S. A. 118, e2014564118. doi:10.1073/pnas.2014564118
Hsieh, C. L., Goldsmith, J. A., Schaub, J. M., Divenere, A. M., Kuo, H. C., Javanmardi, K., et al. (2020). Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505. doi:10.1126/science.abd0826
Hsu, J. C., Laurent-Rolle, M., Pawlak, J. B., Wilen, C. B., and Cresswell, P. (2021). Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein. Proc. Natl. Acad. Sci. U. S. A. 118, e2101161118. doi:10.1073/pnas.2101161118
Hua, C. Z., Miao, Z. P., Zheng, J. S., Huang, Q., Sun, Q. F., Lu, H. P., et al. (2020). Epidemiological features and viral shedding in children with SARS-CoV-2 infection. J. Med. Virol. 92, 2804–2812. doi:10.1002/jmv.26180
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020a). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506. doi:10.1016/s0140-6736(20)30183-5
Huang, C., Wen, T., Shi, F. J., Zeng, X. Y., and Jiao, Y. J. (2020b). Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega 5, 12550–12556. doi:10.1021/acsomega.0c01554
Huang, Y., Yang, C., Xu, X. F., Xu, W., and Liu, S. W. (2020c). Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 41, 1141–1149. doi:10.1038/s41401-020-0485-4
Huang, N., Pérez, P., Kato, T., Mikami, Y., Okuda, K., Gilmore, R. C., et al. (2021). SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 27, 892–903. doi:10.1038/s41591-021-01296-8
Huang, X., Tang, G., Ismail, N., and Wang, X. (2022). Developing RT-LAMP assays for rapid diagnosis of SARS-CoV-2 in saliva. EBioMedicine 75, 103736. doi:10.1016/j.ebiom.2021.103736
Iannaccone, S., Alemanno, F., Houdayer, E., Brugliera, L., Castellazzi, P., Cianflone, D., et al. (2020). COVID-19 rehabilitation units are twice as expensive as regular rehabilitation units. J. Rehabil. Med. 52, jrm00073. doi:10.2340/16501977-2704
Idris, A., Davis, A., Supramaniam, A., Acharya, D., Kelly, G., Tayyar, Y., et al. (2021). A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19. Mol. Ther. 29, 2219–2226. doi:10.1016/j.ymthe.2021.05.004
Ifc (2021). Innovation in manufacturing personal protective equipment: Toward sustainability and circularity. Int. Finance Corp., 1–57.
Ilyas, S., Srivastava, R. R., and Kim, H. (2020). Disinfection technology and strategies for COVID-19 hospital and bio-medical waste management. Sci. Total Environ. 749, 141652. doi:10.1016/j.scitotenv.2020.141652
Ingram, C., Downey, V., Roe, M., Chen, Y., Archibald, M., Kallas, K. A., et al. (2021). COVID-19 prevention and control measures in workplace settings: A rapid review and meta-analysis. Int. J. Environ. Res. Public Health 18, 7847. doi:10.3390/ijerph18157847
International-Committee-on-Taxonomy-of-Viruses (2020). The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5, 536–544. doi:10.1038/s41564-020-0695-z
Ip, D. K., Lau, L. L., Leung, N. H., Fang, V. J., Chan, K. H., Chu, D. K., et al. (2017). Viral shedding and transmission potential of asymptomatic and paucisymptomatic influenza virus infections in the community. Clin. Infect. Dis. 64, 736–742. doi:10.1093/cid/ciw841
Jackson, L. A., Anderson, E. J., Rouphael, N. G., Roberts, P. C., Makhene, M., Coler, R. N., et al. (2020). An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. Overseas. Ed. 383, 1920–1931. doi:10.1056/nejmoa2022483
Jackson, C. B., Farzan, M., Chen, B., and Choe, H. (2022). Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 23, 3–20. doi:10.1038/s41580-021-00418-x
Jacofsky, D., Jacofsky, E. M., and Jacofsky, M. (2020). Understanding antibody testing for COVID-19. J. Arthroplasty 35, S74–s81. doi:10.1016/j.arth.2020.04.055
Jagannathan, P., and Wang, T. T. (2021). Immunity after SARS-CoV-2 infections. Nat. Immunol. 22, 539–540. doi:10.1038/s41590-021-00923-3
Jaimes, J. A., Millet, J. K., and Whittaker, G. R. (2020). Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. iScience 23, 101212. doi:10.1016/j.isci.2020.101212
Jain, V., Duse, A., and Bausch, D. G. (2018). Planning for large epidemics and pandemics: Challenges from a policy perspective. Curr. Opin. Infect. Dis. 31, 316–324. doi:10.1097/qco.0000000000000462
Jairak, W., Chamsai, E., Udom, K., Charoenkul, K., Chaiyawong, S., Techakriengkrai, N., et al. (2022). SARS-CoV-2 delta variant infection in domestic dogs and cats, Thailand. Sci. Rep. 12, 8403. doi:10.1038/s41598-022-12468-y
James, P., Stoddart, D., Harrington, E. D., Beaulaurier, J., Ly, L., Reid, S. W., et al. (2020). LamPORE: Rapid, accurate and highly scalable molecular screening for SARS-CoV-2 infection, based on nanopore sequencing. medRxiv 2020.08.07.20161737. doi:10.1101/2020.08.07.20161737
Jefferson, T., Spencer, E. A., Brassey, J., and Heneghan, C. (2021). Viral cultures for coronavirus disease 2019 infectivity assessment: A systematic review. Clin. Infect. Dis. 73, e3884–e3899. doi:10.1093/cid/ciaa1764
Jeremiah, S. S., Miyakawa, K., Morita, T., Yamaoka, Y., and Ryo, A. (2020). Potent antiviral effect of silver nanoparticles on SARS-CoV-2. Biochem. Biophys. Res. Commun. 533, 195–200. doi:10.1016/j.bbrc.2020.09.018
Jh, V., B, R., S, J., R, S., E, T., B, B., et al. (2020). Personal protective equipment for preventing highly infectious diseases due to exposure to contaminated body fluids in healthcare staff. Cochrane Database Syst. Rev. 5, CD011621. doi:10.1002/14651858.CD011621.pub5
Jiang, J. C., and Zhang, Y. (2020). Serological antibody testing in the COVID-19 pandemic: Their molecular basis and applications. Biochem. Soc. Trans. 48, 2851–2863. doi:10.1042/bst20200744
Jiang, X. L., Zhang, X. L., Zhao, X. N., Li, C. B., Lei, J., Kou, Z. Q., et al. (2020). Transmission potential of asymptomatic and paucisymptomatic severe acute respiratory syndrome coronavirus 2 infections: A 3-family cluster study in China. J. Infect. Dis. 221, 1948–1952. doi:10.1093/infdis/jiaa206
Jin, X., Lian, J. S., Hu, J. H., Gao, J., Zheng, L., Zhang, Y. M., et al. (2020). Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut 69, 1002–1009. doi:10.1136/gutjnl-2020-320926
Johnson, B. A., Xie, X., Kalveram, B., Lokugamage, K. G., Muruato, A., Zou, J., et al. (2020). Furin cleavage site is key to SARS-CoV-2 pathogenesis. bioRxiv 2020.08.26.268854. doi:10.1101/2020.08.26.268854
Johnson, B. A., Xie, X., Bailey, A. L., Kalveram, B., Lokugamage, K. G., Muruato, A., et al. (2021). Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 591, 293–299. doi:10.1038/s41586-021-03237-4
Johnson, T. J., Nishida, R. T., Sonpar, A. P., Lin, Y. J., Watson, K. A., Smith, S. W., et al. (2022). Viral load of SARS-CoV-2 in droplets and bioaerosols directly captured during breathing, speaking and coughing. Sci. Rep. 12, 3484. doi:10.1038/s41598-022-07301-5
Joshi, R. K., Ray, R. K., Adhya, S., Chauhan, V. P. S., and Pani, S. (2021). Spread of COVID-19 by asymptomatic cases: Evidence from military quarantine facilities. BMJ Mil. Health 167, 217–218. doi:10.1136/bmjmilitary-2020-001669
Ju, J. T. J., Boisvert, L. N., and Zuo, Y. Y. (2021). Face masks against COVID-19: Standards, efficacy, testing and decontamination methods. Adv. Colloid Interface Sci. 292, 102435. doi:10.1016/j.cis.2021.102435
Ju, Y., Pu, M., Sun, K., Song, G., and Geng, J. (2022). Nanopore electrochemistry for pathogen detection. Chem. Asian J. 7, e202200774. doi:10.1002/asia.202200774
Jung, J., Rajapakshe, D., Julien, C., and Devaraj, S. (2021). Analytical and clinical performance of cPass neutralizing antibodies assay. Clin. Biochem. 98, 70–73. doi:10.1016/j.clinbiochem.2021.09.008
Jung, J., Lim, Y. J., Kim, E. O., and Kim, S. H. (2022). Risk of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission among healthcare workers dining in hospital staff cafeterias. J. Korean Med. Sci. 37, e14. doi:10.3346/jkms.2022.37.e14
Kadam, V., Truong, Y. B., Schutz, J., Kyratzis, I. L., Padhye, R., and Wang, L. (2021). Gelatin/β–Cyclodextrin Bio–Nanofibers as respiratory filter media for filtration of aerosols and volatile organic compounds at low air resistance. J. Hazard. Mater. 403, 123841. doi:10.1016/j.jhazmat.2020.123841
Kampf, G., Brüggemann, Y., Kaba, H. E. J., Steinmann, J., Pfaender, S., Scheithauer, S., et al. (2020a). Potential sources, modes of transmission and effectiveness of prevention measures against SARS-CoV-2. J. Hosp. Infect. 106, 678–697. doi:10.1016/j.jhin.2020.09.022
Kampf, G., Todt, D., Pfaender, S., and Steinmann, E. (2020b). Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 104, 246–251. doi:10.1016/j.jhin.2020.01.022
Kaname, T., Miyauchi, T., Kuwano, A., Matsuda, Y., Muramatsu, T., and Kajii, T. (1993). Mapping basigin (BSG), a member of the immunoglobulin superfamily, to 19p13.3. Cytogenet. Genome Res. 64, 195–197. doi:10.1159/000133573
Kaneko, N., Kuo, H. H., Boucau, J., Farmer, J. R., Allard-Chamard, H., Mahajan, V. S., et al. (2020). Pillai, S., and Massachusetts consortium on pathogen readiness specimen working, GLoss of bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell 183, 143–157.e13. doi:10.1016/j.cell.2020.08.025
Kang, D., Zuo, X., Yang, R., Xia, F., Plaxco, K. W., and White, R. J. (2009). Comparing the properties of electrochemical-based DNA sensors employing different redox tags. Anal. Chem. 81, 9109–9113. doi:10.1021/ac901811n
Kang, Y. F., Sun, C., Zhuang, Z., Yuan, R. Y., Zheng, Q., Li, J. P., et al. (2021). Rapid development of SARS-CoV-2 spike protein receptor-binding domain self-assembled nanoparticle vaccine candidates. ACS Nano 15, 2738–2752. doi:10.1021/acsnano.0c08379
Kang, K., Chen, Q., Gao, Y., and Yu, K. J. (2022). Detection of SARS-CoV-2 B.1.617.2 (Delta) variant in three cats owned by a confirmed COVID-19 patient in Harbin, China. Vet. Med. Sci. 8, 945–946. doi:10.1002/vms3.715
Kannan, R. M., Nance, E., Kannan, S., and Tomalia, D. A. (2014). Emerging concepts in dendrimer-based nanomedicine: From design principles to clinical applications. J. Intern. Med. 276, 579–617. doi:10.1111/joim.12280
Kantipakala, R., Bonam, S. R., Vemireddy, S., Miryala, S., and Halmuthur, M. S. (2019). Squalane-based emulsion vaccine delivery system: Composition with murabutide activate Th1 response. Pharm. Dev. Technol. 24, 269–275. doi:10.1080/10837450.2018.1469150
Karikó, K., Buckstein, M., Ni, H., and Weissman, D. (2005). Suppression of RNA recognition by toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175. doi:10.1016/j.immuni.2005.06.008
Karim, N., Afroj, S., Lloyd, K., Oaten, L. C., Andreeva, D. V., Carr, C., et al. (2020). Sustainable personal protective clothing for healthcare applications: A review. ACS Nano 14, 12313–12340. doi:10.1021/acsnano.0c05537
Karlsen, T. H. (2022). Understanding COVID-19 through genome-wide association studies. Nat. Genet. 54, 368–369. doi:10.1038/s41588-021-00985-x
Katona, P., Kullar, R., and Zhang, K. (2022). Bringing transmission of SARS-CoV-2 to the surface: Is there a role for fomites? Clin. Infect. Dis. 75, 910–916. doi:10.1093/cid/ciac157
Keech, C., Albert, G., Cho, I., Robertson, A., Reed, P., Neal, S., et al. (2020). Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. Overseas. Ed. 383, 2320–2332. doi:10.1056/nejmoa2026920
Khabazzadeh Tehrani, N., Mahdavi, M., Maleki, F., Zarrati, S., and Tabatabaie, F. (2016). The role of Montanide ISA 70 as an adjuvant in immune responses against Leishmania major induced by thiol-specific antioxidant-based protein vaccine. J. Parasit. Dis. 40, 760–767. doi:10.1007/s12639-014-0574-8
Khademi, F., Taheri, R. A., Momtazi-Borojeni, A. A., Farnoosh, G., Johnston, T. P., and Sahebkar, A. (2018). Potential of cationic liposomes as adjuvants/delivery systems for tuberculosis subunit vaccines. Rev. Physiol. Biochem. Pharmacol. 175, 47–69. doi:10.1007/112_2018_9
Khailany, R. A., Safdar, M., and Ozaslan, M. (2020). Genomic characterization of a novel SARS-CoV-2. Gene Rep. 19, 100682. doi:10.1016/j.genrep.2020.100682
Khanh, N. C., Thai, P. Q., Quach, H. L., Thi, N. H., Dinh, P. C., Duong, T. N., et al. (2020). Transmission of SARS-CoV 2 during long-haul flight. Emerg. Infect. Dis. 26, 2617–2624. doi:10.3201/eid2611.203299
Khare, S., Gurry, C., Freitas, L., Schultz, M. B., Bach, G., Diallo, A., et al. (2021). GISAID's role in pandemic response. China CDC Wkly. 3, 1049–1051. doi:10.46234/ccdcw2021.255
Khoury, D. S., Cromer, D., Reynaldi, A., Schlub, T. E., Wheatley, A. K., Juno, J. A., et al. (2021). Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 27, 1205–1211. doi:10.1038/s41591-021-01377-8
Kianpour, M., Akbarian, M., and Uversky, V. N. (2022). Nanoparticles for coronavirus control. Nanomater. (Basel) 12, 1602. doi:10.3390/nano12091602
Kiesslich, S., and Kamen, A. A. (2020). Vero cell upstream bioprocess development for the production of viral vectors and vaccines. Biotechnol. Adv. 44, 107608. doi:10.1016/j.biotechadv.2020.107608
Kilic, T., Weissleder, R., and Lee, H. (2020). Molecular and immunological diagnostic tests of COVID-19: Current status and challenges. iScience 23, 101406. doi:10.1016/j.isci.2020.101406
Kim, D., Lee, J. Y., Yang, J. S., Kim, J. W., Kim, V. N., and Chang, H. (2020a). The architecture of SARS-CoV-2 transcriptome. Cell 181, 914–921.e10. doi:10.1016/j.cell.2020.04.011
Kim, S. E., Jeong, H. S., Yu, Y., Shin, S. U., Kim, S., Oh, T. H., et al. (2020b). Viral kinetics of SARS-CoV-2 in asymptomatic carriers and presymptomatic patients. Int. J. Infect. Dis. 95, 441–443. doi:10.1016/j.ijid.2020.04.083
Kim, J., Eygeris, Y., Gupta, M., and Sahay, G. (2021a). Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 170, 83–112. doi:10.1016/j.addr.2020.12.014
Kim, Y. I., Kim, D., Yu, K. M., Seo, H. D., Lee, S. A., Casel, M. a. B., et al. (2021b). Development of spike receptor-binding domain nanoparticles as a vaccine candidate against SARS-CoV-2 infection in ferrets. mBio 12 (2), e00230-21. doi:10.1128/mBio.00230-21
Kimball, A., Hatfield, K. M., Arons, M., James, A., Taylor, J., Spicer, K., et al. (2020). Asymptomatic and presymptomatic SARS-CoV-2 infections in residents of a long-term care skilled nursing facility - king county, Washington, march 2020. MMWR. Morb. Mortal. Wkly. Rep. 69, 377–381. doi:10.15585/mmwr.mm6913e1
Kimura, I., Konno, Y., Uriu, K., Hopfensperger, K., Sauter, D., Nakagawa, S., et al. (2021). Sarbecovirus ORF6 proteins hamper induction of interferon signaling. Cell Rep. 34, 108916. doi:10.1016/j.celrep.2021.108916
King, H. a. D., Joyce, M. G., Lakhal-Naouar, I., Ahmed, A., Cincotta, C. M., Subra, C., et al. (2021). Efficacy and breadth of adjuvanted SARS-CoV-2 receptor-binding domain nanoparticle vaccine in macaques. Proc. Natl. Acad. Sci. U. S. A. 118, e2106433118. doi:10.1073/pnas.2106433118
Kirsten, C., Kahre, E., Blankenburg, J., Schumm, L., Haag, L., Galow, L., et al. (2022). Seroprevalence of SARS-CoV-2 in German secondary schools from october 2020 to july 2021: A longitudinal study. Infection 23, 1–8. doi:10.1007/s15010-022-01824-9
Kleanthous, H., Silverman, J. M., Makar, K. W., Yoon, I. K., Jackson, N., and Vaughn, D. W. (2021). Scientific rationale for developing potent RBD-based vaccines targeting COVID-19. NPJ Vaccines 6, 128. doi:10.1038/s41541-021-00393-6
Klebe, L., Felfe, J., and Klug, K. (2021). Healthy leadership in turbulent times: The effectiveness of health-oriented leadership in crisis. Brit. J. Manage. 32, 1203–1218. doi:10.1111/1467-8551.12498
Klinman, D. M., Klaschik, S., Sato, T., and Tross, D. (2009). CpG oligonucleotides as adjuvants for vaccines targeting infectious diseases. Adv. Drug Deliv. Rev. 61, 248–255. doi:10.1016/j.addr.2008.12.012
Klompas, M., Baker, M. A., and Rhee, C. (2020a). Airborne transmission of SARS-CoV-2: Theoretical considerations and available evidence. JAMA 324, 441–442. doi:10.1001/jama.2020.12458
Klompas, M., Morris, C. A., and Shenoy, E. S. (2020b). Universal masking in the covid-19 era. N. Engl. J. Med. Overseas. Ed. 383, e9. doi:10.1056/nejmc2020836
Kluge, H., Martín-Moreno, J. M., Emiroglu, N., Rodier, G., Kelley, E., Vujnovic, M., et al. (2018). Strengthening global health security by embedding the International Health Regulations requirements into national health systems. BMJ Glob. Health 3, e000656. doi:10.1136/bmjgh-2017-000656
Knight, F. C., Gilchuk, P., Kumar, A., Becker, K. W., Sevimli, S., Jacobson, M. E., et al. (2019). Mucosal immunization with a pH-responsive nanoparticle vaccine induces protective CD8(+) lung-resident memory T cells. ACS Nano 13, 10939–10960. doi:10.1021/acsnano.9b00326
Kociolek, L. K., Muller, W. J., Yee, R., Dien Bard, J., Brown, C. A., Revell, P. A., et al. (2020). Comparison of upper respiratory viral load distributions in asymptomatic and symptomatic children diagnosed with SARS-CoV-2 infection in pediatric hospital testing programs. J. Clin. Microbiol. 59 (1), e02593-20. doi:10.1128/JCM.02593-20
Kompaniyets, L., Agathis, N. T., Nelson, J. M., Preston, L. E., Ko, J. Y., Belay, B., et al. (2021). Underlying medical conditions associated with severe COVID-19 illness among children. JAMA Netw. Open 4, e2111182. doi:10.1001/jamanetworkopen.2021.11182
Konda, A., Prakash, A., Moss, G. A., Schmoldt, M., Grant, G. D., and Guha, S. (2020). Aerosol filtration efficiency of common fabrics used in respiratory cloth masks. ACS Nano 14, 6339–6347. doi:10.1021/acsnano.0c03252
Korber, B., Fischer, W. M., Gnanakaran, S., Yoon, H., Theiler, J., Abfalterer, W., et al. (2020). Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 182, 812–827.e19. doi:10.1016/j.cell.2020.06.043
Kräutler, N. J., Suan, D., Butt, D., Bourne, K., Hermes, J. R., Chan, T. D., et al. (2017). Differentiation of germinal center B cells into plasma cells is initiated by high-affinity antigen and completed by Tfh cells. J. Exp. Med. 214, 1259–1267. doi:10.1084/jem.20161533
Kremsner, P. G., Mann, P., Kroidl, A., Leroux-Roels, I., Schindler, C., Gabor, J. J., et al. (2021). Safety and immunogenicity of an mRNA-lipid nanoparticle vaccine candidate against SARS-CoV-2 : A phase 1 randomized clinical trial. Wien. Klin. Wochenschr. 133, 931–941. doi:10.1007/s00508-021-01922-y
Kuhlmann, C., Mayer, C. K., Claassen, M., Maponga, T., Burgers, W. A., Keeton, R., et al. (2022). Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet 399, 625–626. doi:10.1016/s0140-6736(22)00090-3
Kumar, P., FadilaPrasad, A., Akhtar, A., Chaudhary, B. K., Tiwari, L. K., Chaudhry, N., et al. (2021). Vertical transmission and clinical outcome of the neonates born to SARS-CoV-2-positive mothers: A tertiary care hospital-based observational study. BMJ Paediatr. Open 5, e001193. doi:10.1136/bmjpo-2021-001193
Kumar, V., Mishra, S., Sharma, R., Agarwal, J., Ghoshal, U., Khanna, T., et al. (2022). Development of RNA-based assay for rapid detection of SARS-CoV-2 in clinical samples. Intervirology 65, 181–187. doi:10.1159/000522337
Kumari, K., Yadav, R., Mittra, S., Kumar, A., Bajpai, P. K., Srivastava, D. K., et al. (2021). Pregnancy outcomes and vertical transmission capability of SARS-CoV-2 infection among asymptomatic females: A cross-sectional study in a tertiary care rural hospital. J. Fam. Med. Prim. Care 10, 3247–3251. doi:10.4103/jfmpc.jfmpc_23_21
Kuster, G. M., Pfister, O., Burkard, T., Zhou, Q., Twerenbold, R., Haaf, P., et al. (2020). SARS-CoV2: Should inhibitors of the renin-angiotensin system be withdrawn in patients with COVID-19? Eur. Heart J. 41, 1801–1803. doi:10.1093/eurheartj/ehaa235
Kutter, J. S., De Meulder, D., Bestebroer, T. M., Lexmond, P., Mulders, A., Richard, M., et al. (2021). SARS-CoV and SARS-CoV-2 are transmitted through the air between ferrets over more than one meter distance. Nat. Commun. 12, 1653. doi:10.1038/s41467-021-21918-6
Kyle, M. H., Hussain, M., Saltz, V., Mollicone, I., Bence, M., and Dumitriu, D. (2022). Vertical transmission and neonatal outcomes following maternal SARS-CoV-2 infection during pregnancy. Clin. Obstet. Gynecol. 65, 195–202. doi:10.1097/grf.0000000000000667
Kyriakidis, N. C., López-Cortés, A., González, E. V., Grimaldos, A. B., and Prado, E. O. (2021). SARS-CoV-2 vaccines strategies: A comprehensive review of phase 3 candidates. NPJ Vaccines 6, 28. doi:10.1038/s41541-021-00292-w
La Rosa, G., Brandtner, D., Mancini, P., Veneri, C., Bonanno Ferraro, G., Bonadonna, L., et al. (2021). Key SARS-CoV-2 mutations of alpha, gamma, and eta variants detected in urban wastewaters in Italy by long-read amplicon sequencing based on nanopore technology. Water 13, 2503. doi:10.3390/w13182503
Ladhani, S. N., Ireland, G., Baawuah, F., Beckmann, J., Okike, I. O., Ahmad, S., et al. (2022). Emergence of the delta variant and risk of SARS-CoV-2 infection in secondary school students and staff: Prospective surveillance in 18 schools, England. EClinicalMedicine 45, 101319. doi:10.1016/j.eclinm.2022.101319
Laidlaw, B. J., and Ellebedy, A. H. (2022). The germinal centre B cell response to SARS-CoV-2. Nat. Rev. Immunol. 22, 7–18. doi:10.1038/s41577-021-00657-1
Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., et al. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220. doi:10.1038/s41586-020-2180-5
Lau, S. Y., Wang, P., Mok, B. W., Zhang, A. J., Chu, H., Lee, A. C., et al. (2020). Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 9, 837–842. doi:10.1080/22221751.2020.1756700
Le Bert, N., Clapham, H. E., Tan, A. T., Chia, W. N., Tham, C. Y. L., Lim, J. M., et al. (2021). Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 218, e20202617. doi:10.1084/jem.20202617
Le, T. Q. M., Takemura, T., Moi, M. L., Nabeshima, T., Nguyen, L. K. H., Hoang, V. M. P., et al. (2020). Severe acute respiratory syndrome coronavirus 2 shedding by travelers, vietnam, 2020. Emerg. Infect. Dis. 26, 1624–1626. doi:10.3201/eid2607.200591
Leal, L., Pich, J., Ferrer, L., Nava, J., Martí-Lluch, R., Esteban, I., et al. (2022). Safety and immunogenicity of a recombinant protein RBD fusion heterodimer vaccine against SARS-CoV-2: Preliminary results of a phase 1-2a dose-escalating, randomized, double-blind clinical trial. Available at: https://org.medRxiv/abs/200822278560.
Lechien, J. R., Chiesa-Estomba, C. M., De Siati, D. R., Horoi, M., Le Bon, S. D., Rodriguez, A., et al. (2020). Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): A multicenter European study. Eur. Arch. Otorhinolaryngol. 277, 2251–2261. doi:10.1007/s00405-020-05965-1
Lederer, K., Castaño, D., Gómez Atria, D., Oguin, T. H., Wang, S., Manzoni, T. B., et al. (2020). SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity 53, 1281–1295.e5. doi:10.1016/j.immuni.2020.11.009
Ledford, H. (2020). Coronavirus breakthrough: Dexamethasone is first drug shown to save lives. Nature 582, 469. doi:10.1038/d41586-020-01824-5
Lednicky, J. A., Lauzardo, M., Fan, Z. H., Jutla, A., Tilly, T. B., Gangwar, M., et al. (2020). Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients. Int. J. Infect. Dis. 100, 476–482. doi:10.1016/j.ijid.2020.09.025
Lee, E. B., Kim, J. H., Hur, W., Choi, J. E., Kim, S. M., Park, D. J., et al. (2019). Liver-specific gene delivery using engineered virus-like particles of hepatitis E virus. Sci. Rep. 9, 1616. doi:10.1038/s41598-019-38533-7
Lee, I. T., Nakayama, T., Wu, C. T., Goltsev, Y., Jiang, S., Gall, P. A., et al. (2020a). ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat. Commun. 11, 5453. doi:10.1038/s41467-020-19145-6
Lee, S., Kim, T., Lee, E., Lee, C., Kim, H., Rhee, H., et al. (2020b). Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS-CoV-2 infection in a community treatment center in the republic of Korea. JAMA Intern. Med. 180, 1447–1452. doi:10.1001/jamainternmed.2020.3862
Lee, V. J., Aguilera, X., Heymann, D., Wilder-Smith, A., Aguilera, X., et al. (2020c). Preparedness for emerging epidemic threats: A lancet infectious diseases commission. Lancet Infect. Dis. 20, 17–19. doi:10.1016/s1473-3099(19)30674-7
Lee, J., Liu, Z., Chen, W. H., Wei, J., Kundu, R., Adhikari, R., et al. (2021). Process development and scale-up optimization of the SARS-CoV-2 receptor binding domain-based vaccine candidate, RBD219-N1C1. Appl. Microbiol. Biotechnol. 105, 4153–4165. doi:10.1007/s00253-021-11281-3
Leeman, D. S., Ma, T. S., Pathiraja, M. M., Taylor, J. A., Adnan, T. Z., Baltas, I., et al. (2022). Severe acute respiratory coronavirus virus 2 (SARS-CoV-2) nosocomial transmission dynamics, a retrospective cohort study of two healthcare-associated coronavirus disease 2019 (COVID-19) clusters in a district hospital in England during March and April 2020. Infect. Control Hosp. Epidemiol. 43 (11), 1618–1624. doi:10.1017/ice.2021.483
Lei, X., Dong, X., Ma, R., Wang, W., Xiao, X., Tian, Z., et al. (2020). Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 11, 3810. doi:10.1038/s41467-020-17665-9
Lemmin, T., Kalbermatter, D., Harder, D., Plattet, P., and Fotiadis, D. (2020). Structures and dynamics of the novel S1/S2 protease cleavage site loop of the SARS-CoV-2 spike glycoprotein. J. Struct. Biol. X 4, 100038. doi:10.1016/j.yjsbx.2020.100038
Leng, T., Hill, E. M., Holmes, A., Southall, E., Thompson, R. N., Tildesley, M. J., et al. (2022). Quantifying pupil-to-pupil SARS-CoV-2 transmission and the impact of lateral flow testing in English secondary schools. Nat. Commun. 13, 1106. doi:10.1038/s41467-022-28731-9
Lerner, A. M., Folkers, G. K., and Fauci, A. S. (2020). Preventing the spread of SARS-CoV-2 with masks and other "Low-tech" interventions. Jama 324, 1935–1936. doi:10.1001/jama.2020.21946
Lescure, F. X., Bouadma, L., Nguyen, D., Parisey, M., Wicky, P. H., Behillil, S., et al. (2020). Clinical and virological data of the first cases of COVID-19 in europe: A case series. Lancet Infect. Dis. 20, 697–706. doi:10.1016/s1473-3099(20)30200-0
Lessler, J., Grabowski, M. K., Grantz, K. H., Badillo-Goicoechea, E., Metcalf, C. J. E., Lupton-Smith, C., et al. (2021). Household COVID-19 risk and in-person schooling. Science 372, 1092–1097. doi:10.1126/science.abh2939
Leung, A. K., Tam, Y. Y., Chen, S., Hafez, I. M., and Cullis, P. R. (2015a). Microfluidic mixing: A general method for encapsulating macromolecules in lipid nanoparticle systems. J. Phys. Chem. B 119, 8698–8706. doi:10.1021/acs.jpcb.5b02891
Leung, N. H., Xu, C., Ip, D. K., and Cowling, B. J. (2015b). Review article: The fraction of influenza virus infections that are asymptomatic: A systematic review and meta-analysis. Epidemiology 26, 862–872. doi:10.1097/ede.0000000000000340
Leung, C. C., Cheng, K. K., Lam, T. H., and Migliori, G. B. (2020). Mask wearing to complement social distancing and save lives during COVID-19. Int. J. Tuberc. lung Dis. 24, 556–558. doi:10.5588/ijtld.20.0244
Levi, M., Thachil, J., Iba, T., and Levy, J. H. (2020). Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol. 7, e438–e440. doi:10.1016/s2352-3026(20)30145-9
Li, D., and Li, J. (2021). Immunologic testing for SARS-CoV-2 infection from the antigen perspective. J. Clin. Microbiol. 59, 021600–e2220. doi:10.1128/jcm.02160-20
Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., et al. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454. doi:10.1038/nature02145
Li, F., Li, W., Farzan, M., and Harrison, S. C. (2005). Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309, 1864–1868. doi:10.1126/science.1116480
Li, J., Thoon, K. C., Chong, C. Y., Maiwald, M., Kam, K. Q., Nadua, K., et al. (2020a). Comparative analysis of symptomatic and asymptomatic SARS-CoV-2 infection in children. Ann. Acad. Med. Singap. 49, 530–537. doi:10.47102/annals-acadmedsg.2020257
Li, W., Su, Y. Y., Zhi, S. S., Huang, J., Zhuang, C. L., Bai, W. Z., et al. (2020b). Virus shedding dynamics in asymptomatic and mildly symptomatic patients infected with SARS-CoV-2. Clin. Microbiol. Infect. 26, 1556.e1–1556.e6. doi:10.1016/j.cmi.2020.07.008
Li, G., Wang, A., Chen, Y., Sun, Y., Du, Y., Wang, X., et al. (2021a). Development of a colloidal gold-based immunochromatographic strip for rapid detection of severe acute respiratory syndrome coronavirus 2 spike protein. Front. Immunol. 12, 635677. doi:10.3389/fimmu.2021.635677
Li, Y., Liang, M., Gao, L., Ayaz Ahmed, M., Uy, J. P., Cheng, C., et al. (2021b). Face masks to prevent transmission of COVID-19: A systematic review and meta-analysis. Am. J. Infect. Control 49, 900–906. doi:10.1016/j.ajic.2020.12.007
Li, X.-X., Li, C., Du, P.-C., Li, S.-Y., Yu, L., Zhao, Z.-Q., et al. (2022). Rapid and accurate detection of SARS coronavirus 2 by nanopore amplicon sequencing. Front. Microbiol. 13, 735363. doi:10.3389/fmicb.2022.735363
Liefting, L. W., Waite, D. W., and Thompson, J. R. (2021). Application of Oxford nanopore technology to plant virus detection. Viruses 13, 1424. doi:10.3390/v13081424
Lin, X., Bo, W., Guang, Y., and Mario, G. (2012). “Poly(Lactic acid)-based biomaterials: Synthesis, modification and applications,” in Biomedical science, engineering and technology. Editor N. G. Dhanjoo. (Rijeka: IntechOpen), Ch. 11.
Lin, B., Hui, J., and Mao, H. (2021). Nanopore technology and its applications in gene sequencing. Biosens. (Basel) 11, 214. doi:10.3390/bios11070214
Lipsitch, M., Swerdlow, D. L., and Finelli, L. (2020). Defining the epidemiology of covid-19 - studies needed. N. Engl. J. Med. Overseas. Ed. 382, 1194–1196. doi:10.1056/nejmp2002125
Liu, C., Mao, B., Martinez, V., Chen, X., Li, Y., He, L., et al. (2020a). A facile assay for rapid detection of COVID-19 antibodies. RSC Adv. 10, 28041–28048. doi:10.1039/d0ra04107f
Liu, C., Mendonça, L., Yang, Y., Gao, Y., Shen, C., Liu, J., et al. (2020b). The architecture of inactivated SARS-CoV-2 with postfusion spikes revealed by cryo-EM and cryo-ET. Structure 28, 1218–1224. e4e1214. doi:10.1016/j.str.2020.10.001
Liu, C., Yang, Y., Gao, Y., Shen, C., Ju, B., Liu, C., et al. (2020c). Viral architecture of SARS-CoV-2 with post-fusion spike revealed by cryo-EM. bioRxiv, 2020.2003.2002.972927. doi:10.1101/2020.03.02.972927
Liu, W., Zhang, Q., Chen, J., Xiang, R., Song, H., Shu, S., et al. (2020d). Detection of covid-19 in children in early january 2020 in wuhan, China. N. Engl. J. Med. Overseas. Ed. 382, 1370–1371. doi:10.1056/nejmc2003717
Liu, Y., Gayle, A. A., Wilder-Smith, A., and Rocklöv, J. (2020e). The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 27, taaa021. doi:10.1093/jtm/taaa021
Liu, Y., Ning, Z., Chen, Y., Guo, M., Liu, Y., Gali, N. K., et al. (2020f). Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature 582, 557–560. doi:10.1038/s41586-020-2271-3
Liu, D., Wu, F., Cen, Y., Ye, L., Shi, X., Huang, Y., et al. (2021a). Comparative research on nucleocapsid and spike glycoprotein as the rapid immunodetection targets of COVID-19 and establishment of immunoassay strips. Mol. Immunol. 131, 6–12. doi:10.1016/j.molimm.2021.01.005
Liu, L., Kshirsagar, P., Christiansen, J., Gautam, S. K., Aithal, A., Gulati, M., et al. (2021b). Polyanhydride nanoparticles stabilize pancreatic cancer antigen MUC4β. J. Biomed. Mat. Res. A 109, 893–902. doi:10.1002/jbm.a.37080
Liu, Q., Qin, C., Liu, M., and Liu, J. (2021c). Effectiveness and safety of SARS-CoV-2 vaccine in real-world studies: A systematic review and meta-analysis. Infect. Dis. Poverty 10, 132. doi:10.1186/s40249-021-00915-3
Liu, Z., Vanblargan, L. A., Bloyet, L. M., Rothlauf, P. W., Chen, R. E., Stumpf, S., et al. (2021d). Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 29, 477–488. e4e474. doi:10.1016/j.chom.2021.01.014
Logunov, D. Y., Dolzhikova, I. V., Zubkova, O. V., Tukhvatullin, A. I., Shcheblyakov, D. V., Dzharullaeva, A. S., et al. (2020). Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: Two open, non-randomised phase 1/2 studies from Russia. Lancet 396, 887–897. doi:10.1016/s0140-6736(20)31866-3
Logunov, D. Y., Dolzhikova, I. V., Shcheblyakov, D. V., Tukhvatulin, A. I., Zubkova, O. V., Dzharullaeva, A. S., et al. (2021). Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: An interim analysis of a randomised controlled phase 3 trial in Russia. Lancet 397, 671–681. doi:10.1016/s0140-6736(21)00234-8
Lokugamage, K. G., Hage, A., De Vries, M., Valero-Jimenez, A. M., Schindewolf, C., Dittmann, M., et al. (2020). Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 94, e01410–e01420. doi:10.1128/jvi.01410-20
Lokugamage, M. P., Vanover, D., Beyersdorf, J., Hatit, M. Z. C., Rotolo, L., Echeverri, E. S., et al. (2021). Optimization of lipid nanoparticles for the delivery of nebulized therapeutic mRNA to the lungs. Nat. Biomed. Eng. 5, 1059–1068. doi:10.1038/s41551-021-00786-x
Lombardo, D., Kiselev, M. A., and Caccamo, M. T. (2019). Smart nanoparticles for drug delivery application: Development of versatile nanocarrier platforms in Biotechnology and nanomedicine. J. Nanomater. 2019, 1–26. doi:10.1155/2019/3702518
Long, Q.-X., Liu, B.-Z., Deng, H.-J., Wu, G.-C., Deng, K., Chen, Y.-K., et al. (2020). Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 26, 845–848. doi:10.1038/s41591-020-0897-1
Lorenzo-Redondo, R., Nam, H. H., Roberts, S. C., Simons, L. M., Jennings, L. J., Qi, C., et al. (2020). A clade of SARS-CoV-2 viruses associated with lower viral loads in patient upper airways. EBioMedicine 62, 103112. doi:10.1016/j.ebiom.2020.103112
Loucera, C., Perez-Florido, J., Casimiro-Soriguer, C. S., Ortuño, F. M., Carmona, R., Bostelmann, G., et al. (2022). Assessing the impact of SARS-CoV-2 lineages and mutations on patient survival. Viruses 14, 1893. doi:10.3390/v14091893
Løvestad, A. H., Jørgensen, S. B., Handal, N., Ambur, O. H., and Aamot, H. V. (2021). Investigation of intra-hospital SARS-CoV-2 transmission using nanopore whole-genome sequencing. J. Hosp. Infect. 111, 107–116. doi:10.1016/j.jhin.2021.02.022
Low, J. G., De Alwis, R., Chen, S., Kalimuddin, S., Leong, Y. S., Mah, T. K. L., et al. (2021). A phase 1/2 randomized, double-blinded, placebo controlled ascending dose trial to assess the safety, tolerability and immunogenicity of ARCT-021 in healthy adults. medRxiv, 2021.2007.2001.21259831. doi:10.1101/2021.07.01.21259831
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 395, 565–574. doi:10.1016/s0140-6736(20)30251-8
Lua, L. H., Connors, N. K., Sainsbury, F., Chuan, Y. P., Wibowo, N., and Middelberg, A. P. (2014). Bioengineering virus-like particles as vaccines. Biotechnol. Bioeng. 111, 425–440. doi:10.1002/bit.25159
Lubinski, B., Fernandes, M. H. V., Frazier, L., Tang, T., Daniel, S., Diel, D. G., et al. (2022). Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience 25, 103589. doi:10.1016/j.isci.2021.103589
Lucas, C., Klein, J., Sundaram, M., Liu, F., Wong, P., Silva, J., et al. (2020a). Kinetics of antibody responses dictate COVID-19 outcome. medRxiv. doi:10.1101/2020.12.18.20248331
Lucas, C., Wong, P., Klein, J., Castro, T. B. R., Silva, J., Sundaram, M., et al. (2020b). Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584, 463–469. doi:10.1038/s41586-020-2588-y
Lucas, C., Klein, J., Sundaram, M. E., Liu, F., Wong, P., Silva, J., et al. (2021). Delayed production of neutralizing antibodies correlates with fatal COVID-19. Nat. Med. 27, 1178–1186. doi:10.1038/s41591-021-01355-0
Lyngse, F. P., Kirkeby, C. T., Denwood, M., Christiansen, L. E., Mølbak, K., Møller, C. H., et al. (2022). Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: Evidence from Danish households. medRxiv, 2022.2001.2028.22270044. doi:10.1101/2022.01.28.22270044
Ma, X., Zou, F., Yu, F., Li, R., Yuan, Y., Zhang, Y., et al. (2020). Nanoparticle vaccines based on the receptor binding domain (RBD) and Heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity 53, 1315–1330. e9. e1319. doi:10.1016/j.immuni.2020.11.015
Machhi, J., Shahjin, F., Das, S., Patel, M., Abdelmoaty, M. M., Cohen, J. D., et al. (2021). Nanocarrier vaccines for SARS-CoV-2. Adv. Drug Deliv. Rev. 171, 215–239. doi:10.1016/j.addr.2021.01.002
Macintyre, C. R., and Ananda-Rajah, M. R. (2020). Scientific evidence supports aerosol transmission of SARS-COV-2. Antimicrob. Resist. Infect. Control 9, 202. doi:10.1186/s13756-020-00868-6
Madden, E. A., and Diamond, M. S. (2022). Host cell-intrinsic innate immune recognition of SARS-CoV-2. Curr. Opin. Virol. 52, 30–38. doi:10.1016/j.coviro.2021.11.002
Mahapatra, P. S., Chatterjee, S., Tiwari, M. K., Ganguly, R., and Megaridis, C. M. (2020). Surface treatments to enhance the functionality of PPEs. Trans. Indian Natl. Acad. Eng. 5, 333–336. doi:10.1007/s41403-020-00110-0
Mahase, E. (2021). Covid-19: Novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. Bmj 372, n296. doi:10.1136/bmj.n296
Malik, M., Kunze, A. C., Bahmer, T., Herget-Rosenthal, S., and Kunze, T. (2021). SARS-CoV-2: Viral loads of exhaled breath and oronasopharyngeal specimens in hospitalized patients with COVID-19. Int. J. Infect. Dis. 110, 105–110. doi:10.1016/j.ijid.2021.07.012
Malladi, S. K., Patel, U. R., Rajmani, R. S., Singh, R., Pandey, S., Kumar, S., et al. (2021a). Immunogenicity and protective efficacy of a highly thermotolerant, trimeric SARS-CoV-2 receptor binding domain derivative. ACS Infect. Dis. 7, 2546–2564. doi:10.1021/acsinfecdis.1c00276
Malladi, S. K., Singh, R., Pandey, S., Gayathri, S., Kanjo, K., Ahmed, S., et al. (2021b). Design of a highly thermotolerant, immunogenic SARS-CoV-2 spike fragment. J. Biol. Chem. 296, 100025. doi:10.1074/jbc.ra120.016284
Mansbach, R. A., Chakraborty, S., Nguyen, K., Montefiori, D. C., Korber, B., and Gnanakaran, S. (2021). The SARS-CoV-2 Spike variant D614G favors an open conformational state. Sci. Adv. 7, eabf3671. doi:10.1126/sciadv.abf3671
Marcenac, P., Park, G. W., Duca, L. M., Lewis, N. M., Dietrich, E. A., Barclay, L., et al. (2021). Detection of SARS-CoV-2 on surfaces in households of persons with COVID-19. Int. J. Environ. Res. Public Health 18, 8184. doi:10.3390/ijerph18158184
Marchese, V., Formenti, B., Cola, G., Gregori, N., Albini, E., De Palma, G., et al. (2021). Importance of mitigation measures for hospital transmission of SARS-CoV-2 at the onset of the epidemic: The experience of brescia, northern Italy. Infection 49, 1241–1248. doi:10.1007/s15010-021-01692-9
Mardian, Y., Kosasih, H., Karyana, M., Neal, A., and Lau, C. Y. (2021). Review of current COVID-19 diagnostics and opportunities for further development. Front. Med. 8, 615099. doi:10.3389/fmed.2021.615099
Martinez-Flores, D., Zepeda-Cervantes, J., Cruz-Resendiz, A., Aguirre-Sampieri, S., Sampieri, A., and Vaca, L. (2021). SARS-CoV-2 vaccines based on the spike glycoprotein and implications of new viral variants. Front. Immunol. 12, 701501. doi:10.3389/fimmu.2021.701501
Mathew, D., Giles, J. R., Baxter, A. E., Oldridge, D. A., Greenplate, A. R., Wu, J. E., et al. (2020). Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 369, eabc8511. doi:10.1126/science.abc8511
Mauger, D. M., Cabral, B. J., Presnyak, V., Su, S. V., Reid, D. W., Goodman, B., et al. (2019). mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. U. S. A. 116, 24075–24083. doi:10.1073/pnas.1908052116
Maurin, C., He, Z., Mentek, M., Verhoeven, P., Pillet, S., Bourlet, T., et al. (2022). Exploration of the ocular surface infection by SARS-CoV-2 and implications for corneal donation: An ex vivo study. PLoS Med. 19, e1003922. doi:10.1371/journal.pmed.1003922
Mazzoni, A., Salvati, L., Maggi, L., Capone, M., Vanni, A., Spinicci, M., et al. (2020). Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J. Clin. Invest. 130, 4694–4703. doi:10.1172/jci138554
Mcaloon, C., Collins, A., Hunt, K., Barber, A., Byrne, A. W., Butler, F., et al. (2020). Incubation period of COVID-19: A rapid systematic review and meta-analysis of observational research. BMJ Open 10, e039652. doi:10.1136/bmjopen-2020-039652
Mccloskey, B., Dar, O., Zumla, A., and Heymann, D. L. (2014). Emerging infectious diseases and pandemic potential: Status quo and reducing risk of global spread. Lancet Infect. Dis. 14, 1001–1010. doi:10.1016/s1473-3099(14)70846-1
Mckay, P. F., Hu, K., Blakney, A. K., Samnuan, K., Brown, J. C., Penn, R., et al. (2020). Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 11, 3523. doi:10.1038/s41467-020-17409-9
Mcmahan, K., Yu, J., Mercado, N. B., Loos, C., Tostanoski, L. H., Chandrashekar, A., et al. (2021). Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 590, 630–634. doi:10.1038/s41586-020-03041-6
Meinhardt, J., Radke, J., Dittmayer, C., Franz, J., Thomas, C., Mothes, R., et al. (2021). Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 24, 168–175. doi:10.1038/s41593-020-00758-5
Melk, M. M., El-Hawary, S. S., Melek, F. R., Saleh, D. O., Ali, O. M., El Raey, M. A., et al. (2021). Antiviral activity of zinc oxide nanoparticles mediated by plumbago indica L. Extract against herpes simplex virus type 1 (HSV-1). Int. J. Nanomedicine 16, 8221–8233. doi:10.2147/ijn.s339404
Mello, V. M., Eller, C. M., Salvio, A. L., Nascimento, F. F., Figueiredo, C. M., Silva, E., et al. (2022). Effectiveness of face masks in blocking the transmission of SARS-CoV-2: A preliminary evaluation of masks used by SARS-CoV-2-infected individuals. PLoS One 17, e0264389. doi:10.1371/journal.pone.0264389
Mendes, C. R., Dilarri, G., Forsan, C. F., Sapata, V. D. M. R., Lopes, P. R. M., De Moraes, P. B., et al. (2022). Antibacterial action and target mechanisms of zinc oxide nanoparticles against bacterial pathogens. Sci. Rep. 12, 2658. doi:10.1038/s41598-022-06657-y
Menter, T., Haslbauer, J. D., Nienhold, R., Savic, S., Hopfer, H., Deigendesch, N., et al. (2020). Postmortem examination of COVID-19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology 77, 198–209. doi:10.1111/his.14134
Meo, S. A., Bukhari, I. A., Akram, J., Meo, A. S., and Klonoff, D. C. (2021). COVID-19 vaccines: Comparison of biological, pharmacological characteristics and adverse effects of pfizer/BioNTech and Moderna vaccines. Eur. Rev. Med. Pharmacol. Sci. 25, 1663–1669. doi:10.26355/eurrev_202102_24877
Mercado, N. B., Zahn, R., Wegmann, F., Loos, C., Chandrashekar, A., Yu, J., et al. (2020). Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 586, 583–588. doi:10.1038/s41586-020-2607-z
Mertens, P., De Vos, N., Martiny, D., Jassoy, C., Mirazimi, A., Cuypers, L., et al. (2020). Development and potential usefulness of the COVID-19 Ag respi-strip diagnostic assay in a pandemic context. Front. Med. 7, 225. doi:10.3389/fmed.2020.00225
Mertens, J., Coppens, J., Loens, K., Le Mercier, M., Xavier, B. B., Lammens, C., et al. (2022). Monitoring the SARS-CoV-2 pandemic: Screening algorithm with single nucleotide polymorphism detection for the rapid identification of established and emerging variants. Clin. Microbiol. Infect. 28, 124–129. doi:10.1016/j.cmi.2021.09.007
Miles, A. P., Mcclellan, H. A., Rausch, K. M., Zhu, D., Whitmore, M. D., Singh, S., et al. (2005). Montanide ISA 720 vaccines: Quality control of emulsions, stability of formulated antigens, and comparative immunogenicity of vaccine formulations. Vaccine 23, 2530–2539. doi:10.1016/j.vaccine.2004.08.049
Miller, S. M., Cybulski, V., Whitacre, M., Bess, L. S., Livesay, M. T., Walsh, L., et al. (2020). Novel lipidated imidazoquinoline TLR7/8 adjuvants elicit influenza-specific Th1 immune responses and protect against heterologous H3N2 influenza challenge in mice. Front. Immunol. 11, 406. doi:10.3389/fimmu.2020.00406
Min, Y. Q., Huang, M., Sun, X., Deng, F., Wang, H., and Ning, Y. J. (2021). Immune evasion of SARS-CoV-2 from interferon antiviral system. Comput. Struct. Biotechnol. J. 19, 4217–4225. doi:10.1016/j.csbj.2021.07.023
Miorin, L., Kehrer, T., Sanchez-Aparicio, M. T., Zhang, K., Cohen, P., Patel, R. S., et al. (2020). SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U. S. A. 117, 28344–28354. doi:10.1073/pnas.2016650117
Mirjalali, H., Nazemalhosseini-Mojarad, E., Yadegar, A., Mohebbi, S. R., Baghaei, K., Shahrokh, S., et al. (2020). The necessity of stool examination in asymptomatic carriers as a strategic measure to control further spread of SARS-CoV-2. Front. Public Health 8, 553589. doi:10.3389/fpubh.2020.553589
Mitchell, M. J., Billingsley, M. M., Haley, R. M., Wechsler, M. E., Peppas, N. A., and Langer, R. (2021). Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 20, 101–124. doi:10.1038/s41573-020-0090-8
Miyoshi, M., Komagome, R., Yamaguchi, H., Yoshizumi, S., Ishida, S., Nagano, H., et al. (2021). Environmental investigation of SARS-CoV-2 in a karaoke bar: A survey for a cluster of COVID-19 in hokkaido, Japan, 2020. Jpn. J. Infect. Dis. 74, 495–497. doi:10.7883/yoken.jjid.2021.021
Moabelo, K. L., Martin, D. R., Fadaka, A. O., Sibuyi, N. R. S., Meyer, M., and Madiehe, A. M. (2021). Nanotechnology-based strategies for effective and rapid detection of SARS-CoV-2. Mater. (Basel) 14, 7851. doi:10.3390/ma14247851
Moghadas, S. M., Fitzpatrick, M. C., Sah, P., Pandey, A., Shoukat, A., Singer, B. H., et al. (2020). The implications of silent transmission for the control of COVID-19 outbreaks. Proc. Natl. Acad. Sci. U. S. A. 117, 17513–17515. doi:10.1073/pnas.2008373117
Mohsen, M. O., Augusto, G., and Bachmann, M. F. (2020). The 3Ds in virus-like particle based-vaccines: "Design, delivery and dynamics. Immunol. Rev. 296, 155–168. doi:10.1111/imr.12863
Moitra, P., Alafeef, M., Dighe, K., Frieman, M. B., and Pan, D. (2020). Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano 14, 7617–7627. doi:10.1021/acsnano.0c03822
Mokuda, S., Watanabe, H., Kohno, H., Ishitoku, M., Araki, K., Hirata, S., et al. (2022). N(1)-methylpseudouridine-incorporated mRNA enhances exogenous protein expression and suppresses immunogenicity in primary human fibroblast-like synoviocytes. Cytotechnology 74, 503–514. doi:10.1007/s10616-022-00540-4
Mondelli, M. U., Colaneri, M., Seminari, E. M., Baldanti, F., and Bruno, R. (2020). Low risk of SARS-CoV-2 transmission by fomites in real-life conditions. Lancet. Infect. Dis. 21, e112. doi:10.1016/S1473-3099(20)30678-2
Monie, A., Hung, C. F., Roden, R., and Wu, T. C. (2008). Cervarix: A vaccine for the prevention of HPV 16, 18-associated cervical cancer. Biologics. 2, 97–105.
Morais, P., Adachi, H., and Yu, Y. T. (2021). The critical contribution of pseudouridine to mRNA COVID-19 vaccines. Front. Cell Dev. Biol. 9, 789427. doi:10.3389/fcell.2021.789427
Morawska, L., and Cao, J. (2020). Airborne transmission of SARS-CoV-2: The world should face the reality. Environ. Int. 139, 105730. doi:10.1016/j.envint.2020.105730
Moreno-Fierros, L., García-Silva, I., and Rosales-Mendoza, S. (2020). Development of SARS-CoV-2 vaccines: Should we focus on mucosal immunity? Expert Opin. Biol. Ther. 20, 831–836. doi:10.1080/14712598.2020.1767062
Morrison, C. B., Edwards, C. E., Shaffer, K. M., Araba, K. C., Wykoff, J. A., Williams, D. R., et al. (2022). SARS-CoV-2 infection of airway cells causes intense viral and cell shedding, two spreading mechanisms affected by IL-13. Proc. Natl. Acad. Sci. U. S. A. 119, e2119680119. doi:10.1073/pnas.2119680119
Morsli, M., Anani, H., Bréchard, L., Delerce, J., Bedotto, M., Fournier, P. E., et al. (2021). LamPORE SARS-CoV-2 diagnosis and genotyping: A preliminary report. J. Clin. Virol. 138, 104815. doi:10.1016/j.jcv.2021.104815
Moser, C., Amacker, M., and Zurbriggen, R. (2011). Influenza virosomes as a vaccine adjuvant and carrier system. Expert Rev. Vaccines 10, 437–446. doi:10.1586/erv.11.15
Mudgal, R., Nehul, S., and Tomar, S. (2020). Prospects for mucosal vaccine: Shutting the door on SARS-CoV-2. Hum. Vaccin. Immunother. 16, 2921–2931. doi:10.1080/21645515.2020.1805992
Mugglestone, M. A., Ratnaraja, N. V., Bak, A., Islam, J., Wilson, J. A., Bostock, J., et al. (2022). Presymptomatic, asymptomatic and post-symptomatic transmission of SARS-CoV-2: Joint British infection association (BIA), healthcare infection society (HIS), infection prevention society (IPS) and royal College of pathologists (RCPath) guidance. BMC Infect. Dis. 22, 453. doi:10.1186/s12879-022-07440-0
Müller, N. F., Wagner, C., Frazar, C. D., Roychoudhury, P., Lee, J., Moncla, L. H., et al. (2021). Viral genomes reveal patterns of the SARS-CoV-2 outbreak in Washington State. Sci. Transl. Med. 13, eabf0202. doi:10.1126/scitranslmed.abf0202
Muller, C. P. (2021). Do asymptomatic carriers of SARS-COV-2 transmit the virus? Lancet Regional Health - Eur. 4, 100082. doi:10.1016/j.lanepe.2021.100082
Mulligan, M. J., Lyke, K. E., Kitchin, N., Absalon, J., Gurtman, A., Lockhart, S., et al. (2020). Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 586, 589–593. doi:10.1038/s41586-020-2639-4
Munnur, D., Teo, Q., Eggermont, D., Lee, H. H. Y., Thery, F., Ho, J., et al. (2021). Altered ISGylation drives aberrant macrophage-dependent immune responses during SARS-CoV-2 infection. Nat. Immunol. 22, 1416–1427. doi:10.1038/s41590-021-01035-8
Murata, T., Sakurai, A., Suzuki, M., Komoto, S., Ide, T., Ishihara, T., et al. (2021). Shedding of viable virus in asymptomatic SARS-CoV-2 carriers. mSphere 6, e00019–e00021. doi:10.1128/msphere.00019-21
Murewanhema, G., Mukwenha, S., Dzinamarira, T., Mukandavire, Z., Cuadros, D., Madziva, R., et al. (2021). Optimising COVID-19 vaccination policy to mitigate SARS-CoV-2 transmission within schools in Zimbabwe. Vaccines (Basel) 9, 1481. doi:10.3390/vaccines9121481
Murti, M., Achonu, C., Smith, B. T., Brown, K. A., Kim, J. H., Johnson, J., et al. (2021). COVID-19 workplace outbreaks by industry sector and their associated household transmission, ontario, Canada, january to june, 2020. J. Occup. Environ. Med. 63, 574–580. doi:10.1097/jom.0000000000002201
Musa, S. S., Bello, U. M., Zhao, S., Abdullahi, Z. U., Lawan, M. A., and He, D. (2021). Vertical transmission of SARS-CoV-2: A systematic review of systematic reviews. Viruses 13, 1877. doi:10.3390/v13091877
Nagy, A., Horvath, A., Farkas, A., Furi, P., Erdelyi, T., Madas, B. G., et al. (2022). Modeling of nursing care-associated airborne transmission of SARS-CoV-2 in a real-world hospital setting. Geroscience 44, 585–595. doi:10.1007/s11357-021-00512-0
Nakamichi, K., Shen, J. Z., Lee, C. S., Lee, A., Roberts, E. A., Simonson, P. D., et al. (2021). Hospitalization and mortality associated with SARS-CoV-2 viral clades in COVID-19. Sci. Rep. 11, 4802. doi:10.1038/s41598-021-82850-9
Nakayama, T., Lee, I. T., Jiang, S., Matter, M. S., Yan, C. H., Overdevest, J. B., et al. (2021). Determinants of SARS-CoV-2 entry and replication in airway mucosal tissue and susceptibility in smokers. Cell Rep. Med. 2, 100421. doi:10.1016/j.xcrm.2021.100421
Nance, K. D., and Meier, J. L. (2021). Modifications in an emergency: The role of N1-methylpseudouridine in COVID-19 vaccines. ACS Cent. Sci. 7, 748–756. doi:10.1021/acscentsci.1c00197
Nbic+ (2022). Here’s how nanotechnologies tailor face masks for coronavirus era. Available at: https://statnano.com/news/67662/Here%E2%80%99s%20HowNanotechnologies-Tailor-Face-Masks-for-Coronavirus-Era,%20StatNano%20Publication.
Neerukonda, S. N., Vassell, R., Lusvarghi, S., Wang, R., Echegaray, F., Bentley, L., et al. (2021). SARS-CoV-2 Delta variant displays moderate resistance to neutralizing antibodies and spike protein properties of higher soluble ACE2 sensitivity, enhanced cleavage and fusogenic activity. Viruses 13, 2485. doi:10.3390/v13122485
Nelson, J., Sorensen, E. W., Mintri, S., Rabideau, A. E., Zheng, W., Besin, G., et al. (2020). Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 6, eaaz6893. doi:10.1126/sciadv.aaz6893
Ni, W., Yang, X., Yang, D., Bao, J., Li, R., Xiao, Y., et al. (2020). Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care (Houten). 24, 422. doi:10.1186/s13054-020-03120-0
Nickels, L. (2020). Antiviral boost for nanoparticles. Metal. Powder Rep. 75, 330–333. doi:10.1016/j.mprp.2020.10.002
Nicola, M., Alsafi, Z., Sohrabi, C., Kerwan, A., Al-Jabir, A., Iosifidis, C., et al. (2020). The socio-economic implications of the coronavirus pandemic (COVID-19): A review. Int. J. Surg. 78, 185–193. doi:10.1016/j.ijsu.2020.04.018
Niemi, M. E. K., Karjalainen, J., Liao, R. G., Neale, B. M., Daly, M., Ganna, A., et al. (2021). Mapping the human genetic architecture of COVID-19. Nature 600, 472–477. doi:10.1038/s41586-021-03767-x
Nishiyama, N., Kobara, H., Tada, N., Matsui, T., Fujihara, S., Nishiyama, A., et al. (2020). Novel technique using an echo probe cover prevents oral-fecal transmission of SARS-CoV-2 during urgent colonoscopies. Endoscopy 52, E349–e350. doi:10.1055/a-1187-1018
Nissen, K., Krambrich, J., Akaberi, D., Hoffman, T., Ling, J., Lundkvist, A., et al. (2020). Long-distance airborne dispersal of SARS-CoV-2 in COVID-19 wards. Sci. Rep. 10, 19589. doi:10.1038/s41598-020-76442-2
Noh, J. Y., Yoon, J. G., Seong, H., Choi, W. S., Sohn, J. W., Cheong, H. J., et al. (2020). Asymptomatic infection and atypical manifestations of COVID-19: Comparison of viral shedding duration. J. Infect. 81, 816–846. doi:10.1016/j.jinf.2020.05.035
Nooraei, S., Bahrulolum, H., Hoseini, Z. S., Katalani, C., Hajizade, A., Easton, A. J., et al. (2021). Virus-like particles: Preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnology 19, 59. doi:10.1186/s12951-021-00806-7
Notarbartolo, S., Ranzani, V., Bandera, A., Gruarin, P., Bevilacqua, V., Putignano, A. R., et al. (2021). Integrated longitudinal immunophenotypic, transcriptional and repertoire analyses delineate immune responses in COVID-19 patients. Sci. Immunol. 6, eabg5021. doi:10.1126/sciimmunol.abg5021
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28, E63–E63. doi:10.1093/nar/28.12.e63
Nzediegwu, C., and Chang, S. X. (2020). Improper solid waste management increases potential for COVID-19 spread in developing countries. Resour. Conserv. Recycl. 161, 104947. doi:10.1016/j.resconrec.2020.104947
Obermeyer, F., Jankowiak, M., Barkas, N., Schaffner, S. F., Pyle, J. D., Yurkovetskiy, L., et al. (2022). Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness. Science 376, 1327–1332. doi:10.1126/science.abm1208
Oh, S. J., and Shin, O. S. (2022). SARS-CoV-2-mediated evasion strategies for antiviral interferon pathways. J. Microbiol. 60, 290–299. doi:10.1007/s12275-022-1525-1
O'hagan, D. T., Ott, G. S., Nest, G. V., Rappuoli, R., and Giudice, G. D. (2013). The history of MF59(®) adjuvant: A phoenix that arose from the ashes. Expert Rev. Vaccines 12, 13–30. doi:10.1586/erv.12.140
Ohsfeldt, R. L., Choong, C. K., Mc Collam, P. L., Abedtash, H., Kelton, K. A., and Burge, R. (2021). Inpatient hospital costs for COVID-19 patients in the United States. Adv. Ther. 38, 5557–5595. doi:10.1007/s12325-021-01887-4
Olia, A. S., Tsybovsky, Y., Chen, S. J., Liu, C., Nazzari, A. F., Ou, L., et al. (2021). SARS-CoV-2 S2P spike ages through distinct states with altered immunogenicity. J. Biol. Chem. 297, 101127. doi:10.1016/j.jbc.2021.101127
Ong, S. W. X., Lee, P. H., Tan, Y. K., Ling, L. M., Ho, B. C. H., Ng, C. G., et al. (2021). Environmental contamination in a coronavirus disease 2019 (COVID-19) intensive care unit-What is the risk? Infect. Control Hosp. Epidemiol. 42, 669–677. doi:10.1017/ice.2020.1278
Onyeaka, H., Anumudu, C. K., Al-Sharify, Z. T., Egele-Godswill, E., and Mbaegbu, P. (2021). COVID-19 pandemic: A review of the global lockdown and its far-reaching effects. Sci. Prog. 104, 003685042110198. doi:10.1177/00368504211019854
Ortega, R., Gonzalez, M., Nozari, A., and Canelli, R. (2020). Personal protective equipment and covid-19. N. Engl. J. Med. Overseas. Ed. 382, e105. doi:10.1056/nejmvcm2014809
Osan, J. K., Talukdar, S. N., Feldmann, F., Ann Demontigny, B., Jerome, K., Bailey, K. L., et al. (2020). Goblet cell hyperplasia increases SARS-CoV-2 infection in COPD. bioRxiv. doi:10.1101/2020.11.11.379099
Osan, J., Talukdar, S. N., Feldmann, F., Demontigny, B. A., Jerome, K., Bailey, K. L., et al. (2022). Goblet cell hyperplasia increases SARS-CoV-2 infection in chronic obstructive pulmonary disease. Microbiol. Spectr. 10, e0045922. doi:10.1128/spectrum.00459-22
Osipiuk, J., Wydorski, P. M., Lanham, B. T., Tesar, C., Endres, M., Engle, E., et al. (2022). Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin. bioRxiv. doi:10.1101/2021.09.15.460543
O'toole, A., Scher, E., Underwood, A., Jackson, B., Hill, V., Mccrone, J. T., et al. (2021). Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 7, veab064. doi:10.1093/ve/veab064
Ou, T., Mou, H., Zhang, L., Ojha, A., Choe, H., and Farzan, M. (2021). Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog. 17, e1009212. doi:10.1371/journal.ppat.1009212
Oude Munnink, B. B., Sikkema, R. S., Nieuwenhuijse, D. F., Molenaar, R. J., Munger, E., Molenkamp, R., et al. (2021). Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 371, 172–177. doi:10.1126/science.abe5901
Ouyang, Y., Yin, J., Wang, W., Shi, H., Shi, Y., Xu, B., et al. (2020). Downregulated gene expression spectrum and immune responses changed during the disease progression in patients with COVID-19. Clin. Infect. Dis. 71, 2052–2060. doi:10.1093/cid/ciaa462
Overton, E. T., Weir, I. R., Zanni, M. V., Fischinger, S., Macarthur, R. D., Aberg, J. A., et al. (2022). Asymptomatic SARS-CoV-2 infection is common among ART-treated people with HIV. J. Acquir. Immune Defic. Syndr. 90, 377–381. doi:10.1097/QAI.0000000000003000
Owida, H. A., Al-Nabulsi, J. I., Turab, N. M., and Louzi, N. (2022). Nanotechnology role development for COVID-19 pandemic management. J. Nanotechnol. 2022, 1–12. doi:10.1155/2022/1872933
Pachaiappan, R., Rajendran, S., Show, P. L., Manavalan, K., and Naushad, M. (2021). Metal/metal oxide nanocomposites for bactericidal effect: A review. Chemosphere 272, 128607. doi:10.1016/j.chemosphere.2020.128607
Pairo-Castineira, E., Clohisey, S., Klaric, L., Bretherick, A. D., Rawlik, K., Pasko, D., et al. (2021). Genetic mechanisms of critical illness in COVID-19. Nature 591, 92–98. doi:10.1038/s41586-020-03065-y
Papa, G., Mallery, D. L., Albecka, A., Welch, L. G., Cattin-Ortolá, J., Luptak, J., et al. (2021). Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog. 17, e1009246. doi:10.1371/journal.ppat.1009246
Park, J. H., and Lee, H. K. (2021). Delivery routes for COVID-19 vaccines. Vaccines (Basel) 9, 524. doi:10.3390/vaccines9050524
Parr, C. J. C., Wada, S., Kotake, K., Kameda, S., Matsuura, S., Sakashita, S., et al. (2020). N 1-Methylpseudouridine substitution enhances the performance of synthetic mRNA switches in cells. Nucleic Acids Res. 48, e35. doi:10.1093/nar/gkaa070
Parshina-Kottas, Y., Saget, B., Patanjali, K., Fleisher, O., and Gianordoli, G. (2020). This 3-D simulation shows why social distancing is so important. NY Times Available at: https://issues.org/creative-response/3-d-simulation-shows-why-social-distancing-is-so-important/(accessed 14 4 2020).
Passarelli, V. C., Faico-Filho, K., Moreira, L. V. L., Cunha, A. P., Carvalho, J. M. A., Barbosa, G. R., et al. (2021). Asymptomatic COVID-19 in hospital visitors: The underestimated potential of viral shedding. Int. J. Infect. Dis. 102, 412–414. doi:10.1016/j.ijid.2020.10.057
Pastorino, B., Touret, F., Gilles, M., De Lamballerie, X., and Charrel, R. N. (2020). Prolonged infectivity of SARS-CoV-2 in fomites. Emerg. Infect. Dis. 26, 2256–2257. doi:10.3201/eid2609.201788
Patane, L., Cadamuro, M., Massazza, G., Pirola, S., Stagnati, V., Comerio, C., et al. (2022). Evidence of vertical transmission of SARS-CoV-2 and interstitial pneumonia in second-trimester twin stillbirth in asymptomatic woman. Case report and review of the literature. Am. J. Obstet. Gynecol. MFM 4, 100589. doi:10.1016/j.ajogmf.2022.100589
Pater, A. A., Bosmeny, M. S., White, A. A., Sylvain, R. J., Eddington, S. B., Parasrampuria, M., et al. (2021). High throughput nanopore sequencing of SARS-CoV-2 viral genomes from patient samples. J. Biol. Methods 8, e155. doi:10.14440/jbm.2021.360
Patra, J. K., Das, G., Fraceto, L. F., Campos, E. V. R., Rodriguez-Torres, M. D. P., Acosta-Torres, L. S., et al. (2018). Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnology 16, 71. doi:10.1186/s12951-018-0392-8
Paules, C. I., Marston, H. D., and Fauci, A. S. (2020). Coronavirus infections-more than just the common cold. JAMA 323, 707–708. doi:10.1001/jama.2020.0757
Peacock, T. P., Goldhill, D. H., Zhou, J., Baillon, L., Frise, R., Swann, O. C., et al. (2021a). The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 6, 899–909. doi:10.1038/s41564-021-00908-w
Peacock, T. P., Sheppard, C. M., Brown, J. C., Goonawardane, N., Zhou, J., Whiteley, M., et al. (2021b). The SARS-CoV-2 variants associated with infections in India, B.1.617, show enhanced spike cleavage by furin. bioRxiv, 446163. doi:10.1101/2021.05.28.446163
Pecora, A., Malacari, D. A., Mozgovoj, M. V., Diaz, M. L. A., Peralta, A. V., Cacciabue, M., et al. (2022). Anthropogenic infection of domestic cats with SARS-CoV-2 alpha variant B.1.1.7 lineage in buenos aires. Front. Vet. Sci. 9, 790058. doi:10.3389/fvets.2022.790058
Peek, L. J., Middaugh, C. R., and Berkland, C. (2008). Nanotechnology in vaccine delivery. Adv. Drug Deliv. Rev. 60, 915–928. doi:10.1016/j.addr.2007.05.017
Peng, R., Wu, L. A., Wang, Q., Qi, J., and Gao, G. F. (2021a). Cell entry by SARS-CoV-2. Trends biochem. Sci. 46, 848–860. doi:10.1016/j.tibs.2021.06.001
Peng, T., Sui, Z., Huang, Z., Xie, J., Wen, K., Zhang, Y., et al. (2021b). Point-of-care test system for detection of immunoglobulin-G and -M against nucleocapsid protein and spike glycoprotein of SARS-CoV-2. Sensors Actuators B Chem. 331, 129415. doi:10.1016/j.snb.2020.129415
Perri, S., Greer, C. E., Thudium, K., Doe, B., Legg, H., Liu, H., et al. (2003). An alphavirus replicon particle chimera derived from venezuelan equine encephalitis and sindbis viruses is a potent gene-based vaccine delivery vector. J. Virol. 77, 10394–10403. doi:10.1128/jvi.77.19.10394-10403.2003
Perrie, Y., Kastner, E., Khadke, S., Roces, C. B., and Stone, P. (2017). Manufacturing methods for liposome adjuvants. Methods Mol. Biol. 1494, 127–144. doi:10.1007/978-1-4939-6445-1_9
Petersen, E., Wilson, M. E., Touch, S., Mccloskey, B., Mwaba, P., Bates, M., et al. (2016). Rapid spread of Zika virus in the americas--implications for public health preparedness for mass gatherings at the 2016 Brazil olympic games. Int. J. Infect. Dis. 44, 11–15. doi:10.1016/j.ijid.2016.02.001
Petkar, K. C., Patil, S. M., Chavhan, S. S., Kaneko, K., Sawant, K. K., Kunda, N. K., et al. (2021). An overview of nanocarrier-based adjuvants for vaccine delivery. Pharmaceutics 13, 455. doi:10.3390/pharmaceutics13040455
Peto, L., Rodger, G., Carter, D. P., Osman, K. L., Yavuz, M., Johnson, K., et al. (2021). Diagnosis of SARS-CoV-2 infection with LamPORE, a high-throughput platform combining loop-mediated isothermal amplification and nanopore sequencing. J. Clin. Microbiol. 59 (6), e03271-20. doi:10.1128/JCM.03271-20
Pettifor, A., Diprete, B. L., Shook-Sa, B. E., Premkumar, L., Kuczynski, K., Dittmer, D., et al. (2022). A prospective study of asymptomatic SARS-CoV-2 infection among individuals involved in academic research under limited operations during the COVID-19 pandemic. PLoS One 17, e0267353. doi:10.1371/journal.pone.0267353
Pezzini, A., and Padovani, A. (2020). Lifting the mask on neurological manifestations of COVID-19. Nat. Rev. Neurol. 16, 636–644. doi:10.1038/s41582-020-0398-3
Pinelli, F., Perale, G., and Rossi, F. (2020). Coating and functionalization strategies for nanogels and nanoparticles for selective drug delivery. Gels 6, 6. doi:10.3390/gels6010006
Pipolo, C., Bottai, D., Fuccillo, E., Aronica, E., Bruschi, F., Bulfamante, A. M., et al. (2022). Evidence of SARS-CoV-2 in nasal brushings and olfactory mucosa biopsies of COVID-19 patients. PLoS One 17, e0266740. doi:10.1371/journal.pone.0266740
Pisani, A., Pompa, P. P., and Bardi, G. (2020). Potential applications of nanomaterials to quench the cytokine storm in coronavirus disease 19. Front. Bioeng. Biotechnol. 8, 906. doi:10.3389/fbioe.2020.00906
Plante, J. A., Liu, Y., Liu, J., Xia, H., Johnson, B. A., Lokugamage, K. G., et al. (2021). Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592, 116–121. doi:10.1038/s41586-020-2895-3
Plescia, C. B., David, E. A., Patra, D., Sengupta, R., Amiar, S., Su, Y., et al. (2021). SARS-CoV-2 viral budding and entry can be modeled using BSL-2 level virus-like particles. J. Biol. Chem. 296, 100103. doi:10.1074/jbc.ra120.016148
Plummer, E. M., and Manchester, M. (2011). Viral nanoparticles and virus-like particles: Platforms for contemporary vaccine design. WIREs. Nanomed. Nanobiotechnol. 3, 174–196. doi:10.1002/wnan.119
Poggio, C., Colombo, M., Arciola, C. R., Greggi, T., Scribante, A., and Dagna, A. (2020). Copper-alloy surfaces and cleaning regimens against the spread of SARS-CoV-2 in dentistry and orthopedics. From fomites to anti-infective nanocoatings. Mater. (Basel) 13, 3244. doi:10.3390/ma13153244
Polack, F. P., Thomas, S. J., Kitchin, N., Absalon, J., Gurtman, A., Lockhart, S., et al. (2020). Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. Overseas. Ed. 383, 2603–2615. doi:10.1056/nejmoa2034577
Polechova, J., Johnson, K. D., Payne, P., Crozier, A., Beiglbock, M., Plevka, P., et al. (2022). SARS-CoV-2 rapid antigen tests provide benefits for epidemic control - observations from Austrian schools. J. Clin. Epidemiol. 145, 14–19. doi:10.1016/j.jclinepi.2022.01.002
Pollet, J., Chen, W. H., and Strych, U. (2021a). Recombinant protein vaccines, a proven approach against coronavirus pandemics. Adv. Drug Deliv. Rev. 170, 71–82. doi:10.1016/j.addr.2021.01.001
Pollet, J., Chen, W. H., Versteeg, L., Keegan, B., Zhan, B., Wei, J., et al. (2021b). SARS-CoV-2 RBD219-N1C1: A yeast-expressed SARS-CoV-2 recombinant receptor-binding domain candidate vaccine stimulates virus neutralizing antibodies and T-cell immunity in mice. Hum. Vaccin. Immunother. 17, 2356–2366. doi:10.1080/21645515.2021.1901545
Pollock, K. M., Cheeseman, H. M., Szubert, A. J., Libri, V., Boffito, M., Owen, D., et al. (2022). Safety and immunogenicity of a self-amplifying RNA vaccine against COVID-19: COVAC1, a phase I, dose-ranging trial. EClinicalMedicine 44, 101262. doi:10.1016/j.eclinm.2021.101262
Popescu, M., Ştefan, O. M., Ştefan, M., Văleanu, L., and Tomescu, D. (2022). ICU-associated costs during the fourth wave of the COVID-19 pandemic in a tertiary hospital in a low-vaccinated eastern European country. Int. J. Environ. Res. Public Health 19, 1781. doi:10.3390/ijerph19031781
Poudel, A. N., Zhu, S., Cooper, N., Roderick, P., Alwan, N., Tarrant, C., et al. (2021). Impact of covid-19 on health-related quality of life of patients: A structured review. PLoS One 16, e0259164. doi:10.1371/journal.pone.0259164
Prasad, S., Potdar, V., Cherian, S., Abraham, P., Basu, A., and Team, I.-N. N. (2020). Transmission electron microscopy imaging of SARS-CoV-2. Indian J. Med. Res. 151, 241–243. doi:10.4103/ijmr.ijmr_577_20
Prates-Syed, W. A., Chaves, L. C. S., Crema, K. P., Vuitika, L., Lira, A., Côrtes, N., et al. (2021). VLP-based COVID-19 vaccines: An adaptable technology against the threat of new variants. Vaccines (Basel) 9, 1409. doi:10.3390/vaccines9121409
Premkumar, L., Segovia-Chumbez, B., Jadi, R., Martinez, D. R., Raut, R., Markmann, A., et al. (2020). The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 5, eabc8413. doi:10.1126/sciimmunol.abc8413
Promethean-Particles-Ltd (2022). Promethean particles puts its nano-copper to the test in the fight against viruses. Available at: https://prometheanparticles.co.uk/promethean-particles-puts-its-nano-copper-to-the-test-in-the-fight-against-viruses/.
Ptasinska, A., Whalley, C., Bosworth, A., Poxon, C., Bryer, C., Machin, N., et al. (2021). Diagnostic accuracy of loop-mediated isothermal amplification coupled to nanopore sequencing (LamPORE) for the detection of SARS-CoV-2 infection at scale in symptomatic and asymptomatic populations. Clin. Microbiol. Infect. 27, 1348. doi:10.1016/j.cmi.2021.04.008
Puelles, V. G., Lütgehetmann, M., Lindenmeyer, M. T., Sperhake, J. P., Wong, M. N., Allweiss, L., et al. (2020). Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. Overseas. Ed. 383, 590–592. doi:10.1056/nejmc2011400
Qian, C., Liu, X., Xu, Q., Wang, Z., Chen, J., Li, T., et al. (2020a). Recent Progress on the Versatility of Virus-Like Particles, 8. Basel: Vaccines.
Qian, G., Yang, N., Ma, A. H. Y., Wang, L., Li, G., Chen, X., et al. (2020b). COVID-19 transmission within a family cluster by presymptomatic carriers in China. Clin. Infect. Dis. 71, 861–862. doi:10.1093/cid/ciaa316
Qiu, G., Gai, Z., Tao, Y., Schmitt, J., Kullak-Ublick, G. A., and Wang, J. (2020). Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano 14, 5268–5277. doi:10.1021/acsnano.0c02439
Quick, J., Loman, N. J., Duraffour, S., Simpson, J. T., Severi, E., Cowley, L., et al. (2016). Real-time, portable genome sequencing for Ebola surveillance. Nature 530, 228–232. doi:10.1038/nature16996
Quick, J., Grubaugh, N. D., Pullan, S. T., Claro, I. M., Smith, A. D., Gangavarapu, K., et al. (2017). Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 12, 1261–1276. doi:10.1038/nprot.2017.066
Ra, S. H., Lim, J. S., Kim, G. U., Kim, M. J., Jung, J., and Kim, S. H. (2021). Upper respiratory viral load in asymptomatic individuals and mildly symptomatic patients with SARS-CoV-2 infection. Thorax 76, 61–63. doi:10.1136/thoraxjnl-2020-215042
Rajme-Lopez, S., Gonzalez-Lara, M. F., Ortiz-Brizuela, E., Roman-Montes, C. M., Santiago-Cruz, J., Mendoza-Rojas, M. A., et al. (2022). Large-scale screening for severe acute respiratory coronavirus virus 2 (SARS-CoV-2) among healthcare workers: Prevalence and risk factors for asymptomatic and pauci-symptomatic carriers, with emphasis on the use of personal protective equipment (PPE). Infect. Control Hosp. Epidemiol. 43, 513–517. doi:10.1017/ice.2021.68
Raman, R., Patel, K. J., and Ranjan, K. (2021). COVID-19: Unmasking emerging SARS-CoV-2 variants, vaccines and therapeutic strategies. Biomolecules 11, 993. doi:10.3390/biom11070993
Ramasamy, M. N., Minassian, A. M., Ewer, K. J., Flaxman, A. L., Folegatti, P. M., Owens, D. R., et al. (2021). Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): A single-blind, randomised, controlled, phase 2/3 trial. Lancet 396, 1979–1993. doi:10.1016/S0140-6736(20)32466-1
Ramaseshan, R., Sundarrajan, S., Liu, Y., Barhate, R. S., Lala, N. L., and Ramakrishna, S. (2006). Functionalized polymer nanofibre membranes for protection from chemical warfare stimulants. Nanotechnology 17, 2947–2953. doi:10.1088/0957-4484/17/12/021
Rashid, F., Dzakah, E. E., Wang, H., and Tang, S. (2021). The ORF8 protein of SARS-CoV-2 induced endoplasmic reticulum stress and mediated immune evasion by antagonizing production of interferon beta. Virus Res. 296, 198350. doi:10.1016/j.virusres.2021.198350
Rashidzadeh, H., Danafar, H., Rahimi, H., Mozafari, F., Salehiabar, M., Rahmati, M. A., et al. (2021). Nanotechnology against the novel coronavirus (severe acute respiratory syndrome coronavirus 2): Diagnosis, treatment, therapy and future perspectives. Nanomedicine (Lond) 16, 497–516. doi:10.2217/nnm-2020-0441
Rasmi, Y., Saloua, K. S., Nemati, M., and Choi, J. R. (2021). Recent Progress in Nanotechnology for COVID-19 Prevention, Diagnostics and Treatment. Nanomater. (Basel). 11, 1788. doi:10.3390/nano11071788
Rauch, S., Roth, N., Schwendt, K., Fotin-Mleczek, M., Mueller, S. O., and Petsch, B. (2021). mRNA-based SARS-CoV-2 vaccine candidate CVnCoV induces high levels of virus-neutralising antibodies and mediates protection in rodents. NPJ Vaccines 6, 57. doi:10.1038/s41541-021-00311-w
Reddy, S. C., Valderrama, A. L., and Kuhar, D. T. (2019). Improving the use of personal protective equipment: Applying lessons learned. Clin. Infect. Dis. 69, S165–s170. doi:10.1093/cid/ciz619
Regli, A., Sommerfield, A., and Von Ungern-Sternberg, B. S. (2021). The role of fit testing N95/FFP2/FFP3 masks: A narrative review. Anaesthesia 76, 91–100. doi:10.1111/anae.15261
Ren, S. Y., Wang, W. B., Gao, R. D., and Zhou, A. M. (2022). Omicron variant (B.1.1.529) of SARS-CoV-2: Mutation, infectivity, transmission, and vaccine resistance. World J. Clin. Cases 10, 1–11. doi:10.12998/wjcc.v10.i1.1
Reports, W. C. D. C.-S. (2020). Coronavirus disease (COVID-19) weekly epidemiological update and weekly operational update. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/.
Rha, M. S., Jeong, H. W., Ko, J. H., Choi, S. J., Seo, I. H., Lee, J. S., et al. (2021). PD-1-Expressing SARS-CoV-2-specific CD8(+) T cells are not exhausted, but functional in patients with COVID-19. Immunity 54, 44–52.e3. e43. doi:10.1016/j.immuni.2020.12.002
Rios, G., Lacoux, C., Leclercq, V., Diamant, A., Lebrigand, K., Lazuka, A., et al. (2021). Monitoring SARS-CoV-2 variants alterations in Nice neighborhoods by wastewater nanopore sequencing. Lancet Regional Health - Eur. 10, 100202. doi:10.1016/j.lanepe.2021.100202
Rivett, L., Sridhar, S., Sparkes, D., Routledge, M., Jones, N. K., Forrest, S., et al. (2020). Screening of healthcare workers for SARS-CoV-2 highlights the role of asymptomatic carriage in COVID-19 transmission. Elife 9, e58728. doi:10.7554/eLife.58728
Robinot, R., Hubert, M., De Melo, G. D., Lazarini, F., Bruel, T., Smith, N., et al. (2021). SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat. Commun. 12, 4354. doi:10.1038/s41467-021-24521-x
Rodríguez-Ares, T., Lamas-Francis, D., Treviño, M., Navarro, D., Cea, M., López-Valladares, M. J., et al. (2021). SARS-CoV-2 in conjunctiva and tears and ocular symptoms of patients with COVID-19. Vis. (Basel) 5, 51. doi:10.3390/vision5040051
Rohilla, S. (2021). Designing therapeutic strategies to combat severe acute respiratory syndrome coronavirus-2 disease: COVID-19. Drug Dev. Res. 82, 12–26. doi:10.1002/ddr.21720
Rohit, A., Rajasekaran, S., Shenoy, S., Rai, S., Iddya, K., and Dorairajan, S. K. (2021). Reprocessing of N95 masks: Experience from a resource-limited setting in India. Int. J. Infect. Dis. 104, 41–44. doi:10.1016/j.ijid.2020.12.070
Rosenthal, N., Cao, Z., Gundrum, J., Sianis, J., and Safo, S. (2020). Risk factors associated with in-hospital mortality in a US national sample of patients with COVID-19. JAMA Netw. Open 3, e2029058. doi:10.1001/jamanetworkopen.2020.29058
Rosenthal, S. H., Gerasimova, A., Ruiz-Vega, R., Livingston, K., Kagan, R. M., Liu, Y., et al. (2022). Development and validation of a high throughput SARS-CoV-2 whole genome sequencing workflow in a clinical laboratory. Sci. Rep. 12, 2054. doi:10.1038/s41598-022-06091-0
Rothe, C., Schunk, M., Sothmann, P., Bretzel, G., Froeschl, G., Wallrauch, C., et al. (2020). Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. Overseas. Ed. 382, 970–971. doi:10.1056/nejmc2001468
Rowan, N. J., and Moral, R. A. (2021). Disposable face masks and reusable face coverings as non-pharmaceutical interventions (NPIs) to prevent transmission of SARS-CoV-2 variants that cause coronavirus disease (COVID-19): Role of new sustainable NPI design innovations and predictive mathematical modelling. Sci. Total Environ. 772, 145530. doi:10.1016/j.scitotenv.2021.145530
Ruan, Q., Yang, K., Wang, W., Jiang, L., and Song, J. (2020). Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 46, 846–848. doi:10.1007/s00134-020-05991-x
Rydyznski Moderbacher, C., Ramirez, S. I., Dan, J. M., Grifoni, A., Hastie, K. M., Weiskopf, D., et al. (2020). Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 183, 996–1012.e19. doi:10.1016/j.cell.2020.09.038
Sadoff, J., Gray, G., Vandebosch, A., Cárdenas, V., Shukarev, G., Grinsztejn, B., et al. (2021a). Safety and efficacy of single-dose Ad26.COV2.S vaccine against covid-19. N. Engl. J. Med. Overseas. Ed. 384, 2187–2201. doi:10.1056/nejmoa2101544
Sadoff, J., Le Gars, M., Shukarev, G., Heerwegh, D., Truyers, C., De Groot, A. M., et al. (2021b). Interim results of a phase 1-2a trial of Ad26.COV2.S covid-19 vaccine. N. Engl. J. Med. Overseas. Ed. 384, 1824–1835. doi:10.1056/nejmoa2034201
Sáez-Llorens, X., Lanata, C., Aranguren, E., Celis, C. R., Cornejo, R., Deantonio, R., et al. (2022). Safety and immunogenicity of mRNA-LNP COVID-19 vaccine CVnCoV in Latin American adults: A phase 2 randomized study. Vaccine. X 11, 100189. doi:10.1016/j.jvacx.2022.100189
Sah, P., Fitzpatrick, M. C., Zimmer, C. F., Abdollahi, E., Juden-Kelly, L., Moghadas, S. M., et al. (2021). Asymptomatic SARS-CoV-2 infection: A systematic review and meta-analysis. Proc. Natl. Acad. Sci. U. S. A. 118, 17513–17515. doi:10.1073/pnas.2008373117
Saify Nabiabad, H., Amini, M., and Demirdas, S. (2022). Specific delivering of RNAi using spike's aptamer-functionalized lipid nanoparticles for targeting SARS-CoV-2: A strong anti-covid drug in a clinical case study. Chem. Biol. Drug Des. 99, 233–246. doi:10.1111/cbdd.13978
Salamanna, F., Maglio, M., Landini, M. P., and Fini, M. (2020). Body localization of ACE-2: On the trail of the keyhole of SARS-CoV-2. Front. Med. 7, 594495. doi:10.3389/fmed.2020.594495
Salehi-Vaziri, M., Fazlalipour, M., Seyed Khorrami, S. M., Azadmanesh, K., Pouriayevali, M. H., Jalali, T., et al. (2022). The ins and outs of SARS-CoV-2 variants of concern (VOCs). Arch. Virol. 167, 327–344. doi:10.1007/s00705-022-05365-2
Salian, V. S., Wright, J. A., Vedell, P. T., Nair, S., Li, C., Kandimalla, M., et al. (2021). COVID-19 transmission, current treatment, and future therapeutic strategies. Mol. Pharm. 18, 754–771. doi:10.1021/acs.molpharmaceut.0c00608
Salleh, M. Z., Derrick, J. P., and Deris, Z. Z. (2021). Structural evaluation of the spike glycoprotein variants on SARS-CoV-2 transmission and immune evasion. Int. J. Mol. Sci. 22, 7425. doi:10.3390/ijms22147425
Salmenjoki, H., Korhonen, M., Puisto, A., Vuorinen, V., and Alava, M. J. (2021). Modelling aerosol-based exposure to SARS-CoV-2 by an agent based Monte Carlo method: Risk estimates in a shop and bar. PLoS One 16, e0260237. doi:10.1371/journal.pone.0260237
Salvador, F. S., and Fujita, D. M. (2016). Entry routes for Zika virus in Brazil after 2014 world cup: New possibilities. Travel Med. Infect. Dis. 14, 49–51. doi:10.1016/j.tmaid.2015.10.004
Salvador, A., Igartua, M., Hernández, R. M., and Pedraz, J. L. (2011). An overview on the field of micro- and nanotechnologies for synthetic Peptide-based vaccines. J. Drug Deliv. 2011, 1–18. doi:10.1155/2011/181646
Sami, S., Turbyfill, C. R., Daniel-Wayman, S., Shonkwiler, S., Fisher, K. A., Kuhring, M., et al. (2021). Community transmission of SARS-CoV-2 associated with a local bar opening event - Illinois, february 2021. MMWR. Morb. Mortal. Wkly. Rep. 70, 528–532. doi:10.15585/mmwr.mm7014e3
San, J. E., Ngcapu, S., Kanzi, A. M., Tegally, H., Fonseca, V., Giandhari, J., et al. (2021). Transmission dynamics of SARS-CoV-2 within-host diversity in two major hospital outbreaks in South Africa. Virus Evol. 7, veab041. doi:10.1093/ve/veab041
Sanchez, D. G., Gomez Parra, N., Ozden, C., and Rijkers, B. (2020). Which jobs are most vulnerable to COVID-19? What an analysis of the European union reveals. Res. Policy Briefs World Bank Malays. Hub 34, 1–7. doi:10.1596/33737
Santarpia, J. L., Rivera, D. N., Herrera, V. L., Morwitzer, M. J., Creager, H. M., Santarpia, G. W., et al. (2020). Author Correction: Aerosol and surface contamination of SARS-CoV-2 observed in quarantine and isolation care. Sci. Rep. 10, 13892. doi:10.1038/s41598-020-70939-6
Sarfraz, M., Afzal, A., Yang, T., Gai, Y., Raza, S. M., Khan, M. W., et al. (2018). Development of dual drug loaded nanosized liposomal formulation by A reengineered ethanolic injection method and its pre-clinical pharmacokinetic studies. Pharmaceutics 10, 151. doi:10.3390/pharmaceutics10030151
Savoji, M. A., Haghighat, S., Mirzaee, M., Golkaran, B., Mirzaee, R., Esfandiari, B., et al. (2019). Formulation of HBs antigen in Montanide ISA266 shows superiority to commercial HBsAg vaccine in the induction of humoral immune responses. Gastroenterol. Hepatol. Bed Bench 12, 292–300.
Sawatzki, K., Hill, N. J., Puryear, W. B., Foss, A. D., Stone, J. J., and Runstadler, J. A. (2021). Host barriers to SARS-CoV-2 demonstrated by ferrets in a high-exposure domestic setting. Proc. Natl. Acad. Sci. U. S. A. 118, e2025601118. doi:10.1073/pnas.2025601118
Sayers, E. W., Cavanaugh, M., Clark, K., Pruitt, K. D., Schoch, C. L., Sherry, S. T., et al. (2021). GenBank. Nucleic Acids Res. 49, D92–d96. doi:10.1093/nar/gkaa1023
Schoenmaker, L., Witzigmann, D., Kulkarni, J. A., Verbeke, R., Kersten, G., Jiskoot, W., et al. (2021). mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 601, 120586. doi:10.1016/j.ijpharm.2021.120586
Schwendener, R. A. (2014). Liposomes as vaccine delivery systems: A review of the recent advances. Ther. Adv. Vaccines 2, 159–182. doi:10.1177/2051013614541440
Seah, I., and Agrawal, R. (2020). Can the coronavirus disease 2019 (COVID-19) affect the eyes? A review of coronaviruses and ocular implications in humans and animals. Ocul. Immunol. Inflamm. 28, 391–395. doi:10.1080/09273948.2020.1738501
Sedighi, M., Sieber, S., Rahimi, F., Shahbazi, M. A., Rezayan, A. H., Huwyler, J., et al. (2019). Rapid optimization of liposome characteristics using a combined microfluidics and design-of-experiment approach. Drug Deliv. Transl. Res. 9, 404–413. doi:10.1007/s13346-018-0587-4
Seo, G., Lee, G., Kim, M. J., Baek, S. H., Choi, M., Ku, K. B., et al. (2020). Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 14, 5135–5142. doi:10.1021/acsnano.0c02823
Sercombe, L., Veerati, T., Moheimani, F., Wu, S. Y., Sood, A. K., and Hua, S. (2015). Advances and challenges of liposome assisted drug delivery. Front. Pharmacol. 6, 286. doi:10.3389/fphar.2015.00286
Serrano-Aroca, Á., Takayama, K., Tuñón-Molina, A., Seyran, M., Hassan, S. S., Pal Choudhury, P., et al. (2021). Carbon-based nanomaterials: Promising antiviral agents to combat COVID-19 in the microbial-resistant era. ACS Nano 15, 8069–8086. doi:10.1021/acsnano.1c00629
Setti, L., Passarini, F., De Gennaro, G., Barbieri, P., Perrone, M. G., Borelli, M., et al. (2020). Airborne transmission route of COVID-19: Why 2 meters/6 feet of inter-personal distance could not Be enough. Int. J. Environ. Res. Public Health 17, 2932. doi:10.3390/ijerph17082932
Shaffer, L. (2020). 15 drugs being tested to treat COVID-19 and how they would work. Nat. Med. doi:10.1038/d41591-020-00019-9
Shah, R. R., Dodd, S., Schaefer, M., Ugozzoli, M., Singh, M., Otten, G. R., et al. (2015). The development of self-emulsifying oil-in-water emulsion adjuvant and an evaluation of the impact of droplet size on performance. J. Pharm. Sci. 104, 1352–1361. doi:10.1002/jps.24337
Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., et al. (2020a). Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 117, 11727–11734. doi:10.1073/pnas.2003138117
Shang, J., Ye, G., Shi, K., Wan, Y., Luo, C., Aihara, H., et al. (2020b). Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224. doi:10.1038/s41586-020-2179-y
Shang, Y., Li, H., and Zhang, R. (2021). Effects of pandemic outbreak on economies: Evidence from business history context. Front. Public Health 9, 632043. doi:10.3389/fpubh.2021.632043
Shankar, K. (2020). The impact of COVID-19 on IT services industry - expected transformations. Brit. J. Manage. 31, 450–452. doi:10.1111/1467-8551.12423
Sharma, R., Tiwari, S., and Dixit, A. (2021). Covaxin: An overview of its immunogenicity and safety trials in India. Bioinformation 17, 840–845. doi:10.6026/97320630017840
Sheng, J., Amankwah-Amoah, J., Khan, Z., and Wang, X. (2021). COVID-19 pandemic in the new era of big data analytics: Methodological innovations and future research directions. Brit. J. Manage. 32, 1164–1183. doi:10.1111/1467-8551.12441
Shereen, M. A., Khan, S., Kazmi, A., Bashir, N., and Siddique, R. (2020). COVID-19 infection: Emergence, transmission, and characteristics of human coronaviruses. J. Adv. Res. 24, 91–98. doi:10.1016/j.jare.2020.03.005
Shim, B. S., Stadler, K., Nguyen, H. H., Yun, C. H., Kim, D. W., Chang, J., et al. (2012). Sublingual immunization with recombinant adenovirus encoding SARS-CoV spike protein induces systemic and mucosal immunity without redirection of the virus to the brain. Virol. J. 9, 215. doi:10.1186/1743-422x-9-215
Shimabukuro, T.Food and Drug Administration (2021). Allergic reactions including anaphylaxis After receipt of the first dose of pfizer-BioNTech COVID-19 vaccine - United States, december 14-23, 2020. MMWR. Morb. Mortal. Wkly. Rep. 70, 46–51. doi:10.15585/mmwr.mm7002e1
Shin, D., Mukherjee, R., Grewe, D., Bojkova, D., Baek, K., Bhattacharya, A., et al. (2020a). Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 587, 657–662. doi:10.1038/s41586-020-2601-5
Shin, M. D., Shukla, S., Chung, Y. H., Beiss, V., Chan, S. K., Ortega-Rivera, O. A., et al. (2020b). COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 15, 646–655. doi:10.1038/s41565-020-0737-y
Shinde, V., Bhikha, S., Hoosain, Z., Archary, M., Bhorat, Q., Fairlie, L., et al. (2021). Efficacy of NVX-CoV2373 covid-19 vaccine against the B.1.351 variant. N. Engl. J. Med. Overseas. Ed. 384, 1899–1909. doi:10.1056/nejmoa2103055
Shive, M. S., and Anderson, J. M. (1997). Biodegradation and biocompatibility of PLA and PLGA microspheres. Adv. Drug Deliv. Rev. 28, 5–24. doi:10.1016/s0169-409x(97)00048-3
Singanayagam, A., Hakki, S., Dunning, J., Madon, K. J., Crone, M. A., Koycheva, A., et al. (2022). Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: A prospective, longitudinal, cohort study. Lancet Infect. Dis. 22, 183–195. doi:10.1016/s1473-3099(21)00648-4
Singh, R., Burn, J., and Sivan, M. (2020). A letter in response to the SIMFER document on the impact of COVID-19 on Italian rehabilitation services and activities. Eur. J. Phys. Rehabil. Med. 56, 368–369. doi:10.23736/s1973-9087.20.06296-6
Singh, J., Pandit, P., Mcarthur, A. G., Banerjee, A., and Mossman, K. (2021). Evolutionary trajectory of SARS-CoV-2 and emerging variants. Virol. J. 18, 166. doi:10.1186/s12985-021-01633-w
Sivakumar, S. M., Safhi, M. M., Kannadasan, M., and Sukumaran, N. (2011). Vaccine adjuvants - current status and prospects on controlled release adjuvancity. Saudi Pharm. J. 19, 197–206. doi:10.1016/j.jsps.2011.06.003
Sohn, Y., Jeong, S. J., Chung, W. S., Hyun, J. H., Baek, Y. J., Cho, Y., et al. (2020). Assessing viral shedding and infectivity of asymptomatic or mildly symptomatic patients with COVID-19 in a later phase. J. Clin. Med. 9, 2924. doi:10.3390/jcm9092924
Sohrabi, C., Mathew, G., Franchi, T., Kerwan, A., Griffin, M., Soleil, C. D. M. J., et al. (2021). Impact of the coronavirus (COVID-19) pandemic on scientific research and implications for clinical academic training - a review. Int. J. Surg. 86, 57–63. doi:10.1016/j.ijsu.2020.12.008
Sommerstein, R., Fux, C. A., Vuichard-Gysin, D., Abbas, M., Marschall, J., Balmelli, C., et al. (2020). Risk of SARS-CoV-2 transmission by aerosols, the rational use of masks, and protection of healthcare workers from COVID-19. Antimicrob. Resist. Infect. Control 9, 100. doi:10.1186/s13756-020-00763-0
Song, J. W., Zhang, C., Fan, X., Meng, F. P., Xu, Z., Xia, P., et al. (2020). Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 11, 3410. doi:10.1038/s41467-020-17240-2
Song, K. H., Kim, D. M., Lee, H., Ham, S. Y., Oh, S. M., Jeong, H., et al. (2021). Dynamics of viral load and anti-SARS-CoV-2 antibodies in patients with positive RT-PCR results after recovery from COVID-19. Korean J. Intern. Med. 36, 11–14. doi:10.3904/kjim.2020.325
Sonnleitner, S. T., Prelog, M., Sonnleitner, S., Hinterbichler, E., Halbfurter, H., Kopecky, D. B. C., et al. (2022). Cumulative SARS-CoV-2 mutations and corresponding changes in immunity in an immunocompromised patient indicate viral evolution within the host. Nat. Commun. 13, 2560. doi:10.1038/s41467-022-30163-4
Sonovia-Sonomask (2022). Sonovia-Sonomask. Available at: https://sonoviastore.com/
Souri, M., Chiani, M., Farhangi, A., Mehrabi, M. R., Nourouzian, D., Raahemifar, K., et al. (2022). Anti-COVID-19 nanomaterials: Directions to improve prevention, diagnosis, and treatment. Nanomater. (Basel) 12, 783. doi:10.3390/nano12050783
Sportelli, M. C., Izzi, M., Kukushkina, E. A., Hossain, S. I., Picca, R. A., Ditaranto, N., et al. (2020). Can nanotechnology and materials science help the fight against SARS-CoV-2? Nanomater. (Basel) 10, 802. doi:10.3390/nano10040802
Stephenson, K. E., Le Gars, M., Sadoff, J., De Groot, A. M., Heerwegh, D., Truyers, C., et al. (2021). Immunogenicity of the Ad26.COV2.S vaccine for COVID-19. Jama 325, 1535–1544. doi:10.1001/jama.2021.3645
Střížová, Z., Bartůňková, J., and Smrž, D. (2020). Can wearing face masks in public affect transmission route and viral load in COVID-19? Cent. Eur. J. Public Health 28, 161–162. doi:10.21101/cejph.a6290
Stüder, F., Petit, J.-L., Engelen, S., and Mendoza-Parra, M. A. (2021). Real-time SARS-CoV-2 diagnostic and variants tracking over multiple candidates using nanopore DNA sequencing. Sci. Rep. 11, 15869. doi:10.1038/s41598-021-95563-w
Su, Y., Chen, D., Yuan, D., Lausted, C., Choi, J., Dai, C. L., et al. (2020). Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell 183, 1479e1420–1495.e20. doi:10.1016/j.cell.2020.10.037
Sun, L., Zheng, C., and Webster, T. J. (2017). Self-assembled peptide nanomaterials for biomedical applications: Promises and pitfalls. Int. J. Nanomedicine 12, 73–86. doi:10.2147/ijn.s117501
Sun, S., Cai, Y., Song, T. Z., Pu, Y., Cheng, L., Xu, H., et al. (2021). Interferon-armed RBD dimer enhances the immunogenicity of RBD for sterilizing immunity against SARS-CoV-2. Cell Res. 31, 1011–1023. doi:10.1038/s41422-021-00531-8
Sung, A., Bailey, A. L., Stewart, H. B., Mcdonald, D., Wallace, M. A., Peacock, K., et al. (2022). Isolation of SARS-CoV-2 in viral cell culture in immunocompromised patients with persistently positive RT-PCR results. Front. Cell. Infect. Microbiol. 12, 804175. doi:10.3389/fcimb.2022.804175
Surnar, B., Kamran, M. Z., Shah, A. S., and Dhar, S. (2020). Clinically approved antiviral drug in an orally administrable nanoparticle for COVID-19. ACS Pharmacol. Transl. Sci. 3, 1371–1380. doi:10.1021/acsptsci.0c00179
Svitkin, Y. V., Cheng, Y. M., Chakraborty, T., Presnyak, V., John, M., and Sonenberg, N. (2017). N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 45, 6023–6036. doi:10.1093/nar/gkx135
Swain, I. D. (2020). Why the mask? The effectiveness of face masks in preventing the spread of respiratory infections such as COVID-19 - a home testing protocol. J. Med. Eng. Technol. 44, 334–337. doi:10.1080/03091902.2020.1797198
Swanson, P. A., Padilla, M., Hoyland, W., Mcglinchey, K., Fields, P. A., Bibi, S., et al. (2021). AZD1222/ChAdOx1 nCoV-19 vaccination induces a polyfunctional spike protein-specific T(H)1 response with a diverse TCR repertoire. Sci. Transl. Med. 13, eabj7211. doi:10.1126/scitranslmed.abj7211
Tabatabaeizadeh, S. A. (2021). Airborne transmission of COVID-19 and the role of face mask to prevent it: A systematic review and meta-analysis. Eur. J. Med. Res. 26, 1. doi:10.1186/s40001-020-00475-6
Tai, W., Zhang, X., Drelich, A., Shi, J., Hsu, J. C., Luchsinger, L., et al. (2020). A novel receptor-binding domain (RBD)-based mRNA vaccine against SARS-CoV-2. Cell Res. 30, 932–935. doi:10.1038/s41422-020-0387-5
Talebian, S., and Conde, J. (2020). Why go NANO on COVID-19 pandemic? Matter 3, 598–601. doi:10.1016/j.matt.2020.08.005
Tan, F., Wang, K., Liu, J., Liu, D., Luo, J., and Zhou, R. (2020). Viral transmission and clinical features in asymptomatic carriers of SARS-CoV-2 in wuhan, China. Front. Med. 7, 547. doi:10.3389/fmed.2020.00547
Tan, T. K., Rijal, P., Rahikainen, R., Keeble, A. H., Schimanski, L., Hussain, S., et al. (2021). A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nat. Commun. 12, 542. doi:10.1038/s41467-020-20654-7
Tang, T., Jaimes, J. A., Bidon, M. K., Straus, M. R., Daniel, S., and Whittaker, G. R. (2021). Proteolytic activation of SARS-CoV-2 spike at the S1/S2 boundary: Potential role of proteases beyond furin. ACS Infect. Dis. 7, 264–272. doi:10.1021/acsinfecdis.0c00701
Tanriover, M. D., Doğanay, H. L., Akova, M., Güner, H. R., Azap, A., Akhan, S., et al. (2021). Efficacy and safety of an inactivated whole-virion SARS-CoV-2 vaccine (CoronaVac): Interim results of a double-blind, randomised, placebo-controlled, phase 3 trial in Turkey. Lancet 398, 213–222. doi:10.1016/s0140-6736(21)01429-x
Tao, K., Tzou, P. L., Nouhin, J., Gupta, R. K., De Oliveira, T., Kosakovsky Pond, S. L., et al. (2021). The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 22, 757–773. doi:10.1038/s41576-021-00408-x
Tatsis, N., and Ertl, H. C. (2004). Adenoviruses as vaccine vectors. Mol. Ther. 10, 616–629. doi:10.1016/j.ymthe.2004.07.013
Teijaro, J. R., and Farber, D. L. (2021). COVID-19 vaccines: Modes of immune activation and future challenges. Nat. Rev. Immunol. 21, 195–197. doi:10.1038/s41577-021-00526-x
Tenchov, R., Bird, R., Curtze, A. E., and Zhou, Q. (2021). Lipid nanoparticles-from liposomes to mRNA vaccine delivery, a landscape of research diversity and advancement. ACS Nano 15, 16982–17015. doi:10.1021/acsnano.1c04996
The-Covid-Host-Genetics-Initiative (2020). The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur. J. Hum. Genet. 28, 715–718. doi:10.1038/s41431-020-0636-6
Thompson, M. G., Burgess, J. L., Naleway, A. L., Tyner, H., Yoon, S. K., Meece, J., et al. (2021). Prevention and attenuation of covid-19 with the BNT162b2 and mRNA-1273 vaccines. N. Engl. J. Med. Overseas. Ed. 385, 320–329. doi:10.1056/nejmoa2107058
Tian, B., Gao, F., Fock, J., Dufva, M., and Hansen, M. F. (2020). Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. X. 165, 112356. doi:10.1016/j.bios.2020.112356
Tian, J. H., Patel, N., Haupt, R., Zhou, H., Weston, S., Hammond, H., et al. (2021). SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat. Commun. 12, 372. doi:10.1038/s41467-020-20653-8
Tiboni, M., Casettari, L., and Illum, L. (2021). Nasal vaccination against SARS-CoV-2: Synergistic or alternative to intramuscular vaccines? Int. J. Pharm. X. 603, 120686. doi:10.1016/j.ijpharm.2021.120686
Tidu, A., Janvier, A., Schaeffer, L., Sosnowski, P., Kuhn, L., Hammann, P., et al. (2020). The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. Rna 27, 253–264. doi:10.1261/rna.078121.120
Tikellis, C., and Thomas, M. C. (2012). Angiotensin-converting enzyme 2 (ACE2) is a key modulator of the renin angiotensin system in health and disease. Int. J. Pept. 2012, 256294. doi:10.1155/2012/256294
Tinkle, S., Mcneil, S. E., Mühlebach, S., Bawa, R., Borchard, G., Barenholz, Y. C., et al. (2014). Nanomedicines: Addressing the scientific and regulatory gap. Ann. N. Y. Acad. Sci. 1313, 35–56. doi:10.1111/nyas.12403
To, K. K., Hung, I. F., Ip, J. D., Chu, A. W., Chan, W. M., Tam, A. R., et al. (2021). Coronavirus disease 2019 (COVID-19) Re-infection by a phylogenetically distinct severe acute respiratory syndrome coronavirus 2 strain confirmed by whole genome sequencing. Clin. Infect. Dis. 73, e2946–e2951. doi:10.1093/cid/ciaa1275
Toback, S., Galiza, E., Cosgrove, C., Galloway, J., Goodman, A. L., Swift, P. A., et al. (2022). Safety, immunogenicity, and efficacy of a COVID-19 vaccine (NVX-CoV2373) co-administered with seasonal influenza vaccines: An exploratory substudy of a randomised, observer-blinded, placebo-controlled, phase 3 trial. Lancet. Respir. Med. 10, 167–179. doi:10.1016/S2213-2600(21)00409-4
Toscano, G., Palmerini, F., Ravaglia, S., Ruiz, L., Invernizzi, P., Cuzzoni, M. G., et al. (2020). Guillain-barré syndrome associated with SARS-CoV-2. N. Engl. J. Med. Overseas. Ed. 382, 2574–2576. doi:10.1056/nejmc2009191
Tsivgoulis, G., Palaiodimou, L., Katsanos, A. H., Caso, V., Köhrmann, M., Molina, C., et al. (2020). Neurological manifestations and implications of COVID-19 pandemic. Ther. Adv. Neurol. Disord. 13, 175628642093203. doi:10.1177/1756286420932036
Tuaillon, E., Bolloré, K., Pisoni, A., Debiesse, S., Renault, C., Marie, S., et al. (2020). Detection of SARS-CoV-2 antibodies using commercial assays and seroconversion patterns in hospitalized patients. J. Infect. 81, e39–e45. doi:10.1016/j.jinf.2020.05.077
Turner, J. S., O'halloran, J. A., Kalaidina, E., Kim, W., Schmitz, A. J., Zhou, J. Q., et al. (2021). SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature 596, 109–113. doi:10.1038/s41586-021-03738-2
Uhm, J. S., Ahn, J. Y., Hyun, J., Sohn, Y., Kim, J. H., Jeong, S. J., et al. (2020). Patterns of viral clearance in the natural course of asymptomatic COVID-19: Comparison with symptomatic non-severe COVID-19. Int. J. Infect. Dis. 99, 279–285. doi:10.1016/j.ijid.2020.07.070
Underwood, B. A., Yankie, L., Nawrocki, E. P., Palanigobu, V., Gotvyanskyy, S., Calhoun, V. C., et al. (2022). Rapid automated validation, annotation and publication of SARS-CoV-2 sequences to GenBank. Database (Oxford). 2022, baac006. doi:10.1093/database/baac006
Vacca, D., Fiannaca, A., Tramuto, F., Cancila, V., La Paglia, L., Mazzucco, W., et al. (2022). Direct RNA nanopore sequencing of SARS-CoV-2 extracted from critical material from swabs. Life (Basel) 12, 69. doi:10.3390/life12010069
Vahedifard, F., and Chakravarthy, K. (2021). Nanomedicine for COVID-19: The role of nanotechnology in the treatment and diagnosis of COVID-19. emergent Mat. 4, 75–99. doi:10.1007/s42247-021-00168-8
Valcourt, D. M., Dang, M. N., Scully, M. A., and Day, E. S. (2020). Nanoparticle-Mediated Co-delivery of notch-1 antibodies and ABT-737 as a potent treatment strategy for triple-negative breast cancer. ACS Nano 14, 3378–3388. doi:10.1021/acsnano.9b09263
Valdez-Salas, B., Beltran-Partida, E., Cheng, N., Salvador-Carlos, J., Valdez-Salas, E. A., Curiel-Alvarez, M., et al. (2021). Promotion of surgical masks antimicrobial activity by disinfection and impregnation with disinfectant silver nanoparticles. Int. J. Nanomedicine 16, 2689–2702. doi:10.2147/ijn.s301212
Valencak, T. G., Csiszar, A., Szalai, G., Podlutsky, A., Tarantini, S., Fazekas-Pongor, V., et al. (2021). Animal reservoirs of SARS-CoV-2: Calculable COVID-19 risk for older adults from animal to human transmission. Geroscience 43, 2305–2320. doi:10.1007/s11357-021-00444-9
Valera, E., Jankelow, A., Lim, J., Kindratenko, V., Ganguli, A., White, K., et al. (2021). COVID-19 point-of-care diagnostics: Present and future. ACS Nano 15, 7899–7906. doi:10.1021/acsnano.1c02981
Van Der Jeught, K., De Koker, S., Bialkowski, L., Heirman, C., Tjok Joe, P., Perche, F., et al. (2018). Dendritic cell targeting mRNA lipopolyplexes combine strong antitumor T-cell immunity with improved inflammatory safety. ACS Nano 12, 9815–9829. doi:10.1021/acsnano.8b00966
Van Dijk, E. L., Jaszczyszyn, Y., Naquin, D., and Thermes, C. (2018). The third revolution in sequencing technology. Trends Genet. 34, 666–681. doi:10.1016/j.tig.2018.05.008
Van Dongen, J. E., Berendsen, J. T. W., Steenbergen, R. D. M., Wolthuis, R. M. F., Eijkel, J. C. T., and Segerink, L. I. (2020). Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities. Biosens. Bioelectron. X. 166, 112445. doi:10.1016/j.bios.2020.112445
Van Doorn, E., Liu, H., Huckriede, A., and Hak, E. (2016). Safety and tolerability evaluation of the use of montanide ISA™51 as vaccine adjuvant: A systematic review. Hum. Vaccin. Immunother. 12, 159–169. doi:10.1080/21645515.2015.1071455
Van Doremalen, N., Bushmaker, T., Morris, D. H., Holbrook, M. G., Gamble, A., Williamson, B. N., et al. (2020). Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. Overseas. Ed. 382, 1564–1567. doi:10.1056/nejmc2004973
Vandenberg, O., Martiny, D., Rochas, O., Van Belkum, A., and Kozlakidis, Z. (2021). Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 19, 171–183. doi:10.1038/s41579-020-00461-z
Varahachalam, S. P., Lahooti, B., Chamaneh, M., Bagchi, S., Chhibber, T., Morris, K., et al. (2021). Nanomedicine for the SARS-CoV-2: State-of-the-Art and future prospects. Int. J. Nanomedicine 16, 539–560. doi:10.2147/ijn.s283686
Varypataki, E. M., Van Der Maaden, K., Bouwstra, J., Ossendorp, F., and Jiskoot, W. (2015). Cationic liposomes loaded with a synthetic long peptide and poly(I:C): A defined adjuvanted vaccine for induction of antigen-specific T cell cytotoxicity. Aaps J. 17, 216–226. doi:10.1208/s12248-014-9686-4
Vazquez, C., Swanson, S. E., Negatu, S. G., Dittmar, M., Miller, J., Ramage, H. R., et al. (2021). SARS-CoV-2 viral proteins NSP1 and NSP13 inhibit interferon activation through distinct mechanisms. PLoS One 16, e0253089. doi:10.1371/journal.pone.0253089
Verbeke, R., Lentacker, I., De Smedt, S. C., and Dewitte, H. (2021). The dawn of mRNA vaccines: The COVID-19 case. J. Control. Release 333, 511–520. doi:10.1016/j.jconrel.2021.03.043
Verbeke, A. (2020). Will the COVID-19 pandemic really change the governance of global value chains? Brit. J. Manage. 31, 444–446. doi:10.1111/1467-8551.12422
Verdoni, L., Mazza, A., Gervasoni, A., Martelli, L., Ruggeri, M., Ciuffreda, M., et al. (2020). An outbreak of severe kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: An observational cohort study. Lancet 395, 1771–1778. doi:10.1016/s0140-6736(20)31103-x
Viehweger, A., Krautwurst, S., Lamkiewicz, K., Madhugiri, R., Ziebuhr, J., Hölzer, M., et al. (2019). Direct RNA nanopore sequencing of full-length coronavirus genomes provides novel insights into structural variants and enables modification analysis. Genome Res. 29, 1545–1554. doi:10.1101/gr.247064.118
Villoutreix, B. O., Badiola, I., and Khatib, A.-M. (2022). Furin and COVID-19: Structure, function and chemoinformatic analysis of representative active site inhibitors. Front. Drug Discov. (Lausanne). 2. doi:10.3389/fddsv.2022.899239
Viner, R., Waddington, C., Mytton, O., Booy, R., Cruz, J., Ward, J., et al. (2022). Transmission of SARS-CoV-2 by children and young people in households and schools: A meta-analysis of population-based and contact-tracing studies. J. Infect. 84, 361–382. doi:10.1016/j.jinf.2021.12.026
Vitiello, A., Ferrara, F., Troiano, V., and La Porta, R. (2021). COVID-19 vaccines and decreased transmission of SARS-CoV-2. Inflammopharmacology 29, 1357–1360. doi:10.1007/s10787-021-00847-2
Vogel, A. B., Lambert, L., Kinnear, E., Busse, D., Erbar, S., Reuter, K. C., et al. (2018). Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 26, 446–455. doi:10.1016/j.ymthe.2017.11.017
Volpatti, L. R., Matranga, M. A., Cortinas, A. B., Delcassian, D., Daniel, K. B., Langer, R., et al. (2020). Glucose-responsive nanoparticles for rapid and extended self-regulated insulin delivery. ACS Nano 14, 488–497. doi:10.1021/acsnano.9b06395
Volz, E., Mishra, S., Chand, M., Barrett, J. C., Johnson, R., Geidelberg, L., et al. (2021). Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 593, 266–269. doi:10.1038/s41586-021-03470-x
Von Wintersdorff, C. J. H., Dingemans, J., Van Alphen, L. B., Wolffs, P. F. G., Van Der Veer, B. M. J. W., Hoebe, C. J. P. A., et al. (2022). Infections with the SARS-CoV-2 Delta variant exhibit fourfold increased viral loads in the upper airways compared to Alpha or non-variants of concern. Sci. Rep. 12, 13922. doi:10.1038/s41598-022-18279-5
Voskarides, K. (2020). Animal-to-Human viral transitions: Is SARS-CoV-2 an evolutionarily successful one? J. Mol. Evol. 88, 421–423. doi:10.1007/s00239-020-09947-z
Voysey, M., Clemens, S. a. C., Madhi, S. A., Weckx, L. Y., Folegatti, P. M., Aley, P. K., et al. (2021). Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, south Africa, and the UK. Lancet 397, 99–111. doi:10.1016/S0140-6736(20)32661-1
Wafa, E. I., Geary, S. M., Goodman, J. T., Narasimhan, B., and Salem, A. K. (2017). The effect of polyanhydride chemistry in particle-based cancer vaccines on the magnitude of the anti-tumor immune response. Acta Biomater. 50, 417–427. doi:10.1016/j.actbio.2017.01.005
Wake, A. D. (2022). Intrauterine vertical transmission of SARS-CoV-2 infection among confirmed cases of pregnant women: "A double burden for the pregnant women"-A systematic review. Glob. Pediatr. Health 9, 2333794X2210897. doi:10.1177/2333794x221089765
Walls, A. C., Fiala, B., Schäfer, A., Wrenn, S., Pham, M. N., Murphy, M., et al. (2020a). Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 183, 1367–1382.e17. e1317. doi:10.1016/j.cell.2020.10.043
Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., Mcguire, A. T., and Veesler, D. (2020b). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 181, 1735–2292. e286. doi:10.1016/j.cell.2020.11.032
Walsh, E. E., Frenck, R., Falsey, A. R., Kitchin, N., Absalon, J., Gurtman, A., et al. (2020a). RNA-based COVID-19 vaccine BNT162b2 selected for a pivotal efficacy study. medRxiv., 2020.20176651. doi:10.1101/2020.08.17.20176651
Walsh, E. E., Frenck, R. W., Falsey, A. R., Kitchin, N., Absalon, J., Gurtman, A., et al. (2020b). Safety and immunogenicity of two RNA-based covid-19 vaccine candidates. N. Engl. J. Med. Overseas. Ed. 383, 2439–2450. doi:10.1056/nejmoa2027906
Wan, Y., Shang, J., Graham, R., Baric, R. S., and Li, F. (2020). Receptor recognition by the novel coronavirus from wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 94, 001277. doi:10.1128/jvi.00127-20
Wang, F., Kream, R. M., and Stefano, G. B. (2020a). An evidence based perspective on mRNA-SARS-CoV-2 vaccine development. Med. Sci. Monit. 26, e924700. doi:10.12659/msm.924700
Wang, H., Zhang, Y., Huang, B., Deng, W., Quan, Y., Wang, W., et al. (2020b). Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 182, 713–721.e9. e719. doi:10.1016/j.cell.2020.06.008
Wang, K., Chen, W., Zhang, Z., Deng, Y., Lian, J. Q., Du, P., et al. (2020c). CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 5, 283. doi:10.1038/s41392-020-00426-x
Wang, M. Y., Zhao, R., Gao, L. J., Gao, X. F., Wang, D. P., and Cao, J. M. (2020d). SARS-CoV-2: Structure, biology, and structure-based therapeutics development. Front. Cell. Infect. Microbiol. 10, 6. doi:10.3389/fcimb.2020.00006
Wang, Q., Zhang, Y., Wu, L., Niu, S., Song, C., Zhang, Z., et al. (2020e). Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 181, 894–904.e9. e899. doi:10.1016/j.cell.2020.03.045
Wang, W., Xu, Y., Gao, R., Lu, R., Han, K., Wu, G., et al. (2020f). Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323, 1843–1844. doi:10.1001/jama.2020.3786
Wang, Y., He, Y., Tong, J., Qin, Y., Xie, T., Li, J., et al. (2020g). Characterization of an asymptomatic cohort of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infected individuals outside of wuhan, China. Clin. Infect. Dis. 71, 2132–2138. doi:10.1093/cid/ciaa629
Wang, D., Zhang, B., Ding, H., Liu, D., Xiang, J., Gao, X. J., et al. (2021a). TiO2 supported single Ag atoms nanozyme for elimination of SARS-CoV2. Nano Today 40, 101243. doi:10.1016/j.nantod.2021.101243
Wang, E. Y., Mao, T., Klein, J., Dai, Y., Huck, J. D., Jaycox, J. R., et al. (2021b). Diverse functional autoantibodies in patients with COVID-19. Nature 595, 283–288. doi:10.1038/s41586-021-03631-y
Wang, R., Yang, X., Chang, M., Xue, Z., Wang, W., Bai, L., et al. (2021c). ORF3a protein of severe acute respiratory syndrome coronavirus 2 inhibits interferon-activated janus kinase/signal transducer and activator of transcription signaling via elevating suppressor of cytokine signaling 1. Front. Microbiol. 12, 752597. doi:10.3389/fmicb.2021.752597
Wang, W., Zhou, Z., Xiao, X., Tian, Z., Dong, X., Wang, C., et al. (2021d). SARS-CoV-2 nsp12 attenuates type I interferon production by inhibiting IRF3 nuclear translocation. Cell. Mol. Immunol. 18, 945–953. doi:10.1038/s41423-020-00619-y
Wang, Y., Zhao, Y., Bollas, A., Wang, Y., and Au, K. F. (2021e). Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 39, 1348–1365. doi:10.1038/s41587-021-01108-x
Wang, Z., Lorenzi, J. C. C., Muecksch, F., Finkin, S., Viant, C., Gaebler, C., et al. (2021f). Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 13, eabf1555. doi:10.1126/scitranslmed.abf1555
Wang, R., Chen, J., Hozumi, Y., Yin, C., and Wei, G. W. (2022a). Emerging vaccine-breakthrough SARS-CoV-2 variants. ACS Infect. Dis. 8, 546–556. doi:10.1021/acsinfecdis.1c00557
Wang, Y., Chen, D., Zhu, C., Zhao, Z., Gao, S., Gou, J., et al. (2022b). Genetic surveillance of five SARS-CoV-2 clinical samples in henan province using nanopore sequencing. Front. Immunol. 13, 814806. doi:10.3389/fimmu.2022.814806
Wang, Y., Liu, C., Zhang, C., Wang, Y., Hong, Q., Xu, S., et al. (2022c). Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies. Nat. Commun. 13, 871. doi:10.1038/s41467-022-28528-w
Wawina-Bokalanga, T., Logist, A. S., Sinnesael, R., Van Holm, B., Delforge, M. L., Struyven, P., et al. (2022). Genomic evidence of co-identification with omicron and delta SARS-CoV-2 variants: A report of two cases. Int. J. Infect. Dis. 122, 212–214. doi:10.1016/j.ijid.2022.05.043
Wei, C., Wan, L., Yan, Q., Wang, X., Zhang, J., Yang, X., et al. (2020). HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. Metab. 2, 1391–1400. doi:10.1038/s42255-020-00324-0
Weisblum, Y., Schmidt, F., Zhang, F., Dasilva, J., Poston, D., Lorenzi, J. C., et al. (2020). Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 9, e61312. doi:10.7554/elife.61312
Weiss, C., Carriere, M., Fusco, L., Capua, I., Regla-Nava, J. A., Pasquali, M., et al. (2020). Toward nanotechnology-enabled approaches against the COVID-19 pandemic. ACS Nano 14, 6383–6406. doi:10.1021/acsnano.0c03697
White, L. F., Murray, E. J., and Chakravarty, A. (2022). The role of schools in driving SARS-CoV-2 transmission: Not just an open-and-shut case. Cell. Rep. Med. 3, 100556. doi:10.1016/j.xcrm.2022.100556
Whittaker, G. R. (2021). SARS-CoV-2 spike and its adaptable furin cleavage site. Lancet Microbe 2, e488–e489. doi:10.1016/s2666-5247(21)00174-9
WHO (1987). Acceptability of cell substrates for production of biologicals. Report of a WHO Study Group on Biologicals. World Health Organ. Tech. Rep. Ser. 747, 1–29.
WHO (2022). Modes of transmission of virus causing COVID-19: Implications for IPC precaution recommendations. Available at: https://www.who.int/news-room/commentaries/detail/modes-of-transmission-of-virus-causing-covid-19-implications- for-ipc-precaution-recommendations (accessed on January 1, 2022).
Widge, A. T., Rouphael, N. G., Jackson, L. A., Anderson, E. J., Roberts, P. C., Makhene, M., et al. (2021). Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N. Engl. J. Med. Overseas. Ed. 384, 80–82. doi:10.1056/nejmc2032195
Wilmes, P., Zimmer, J., Schulz, J., Glod, F., Veiber, L., Mombaerts, L., et al. (2021). SARS-CoV-2 transmission risk from asymptomatic carriers: Results from a mass screening programme in Luxembourg. Lancet Regional Health - Eur. 4, 100056. doi:10.1016/j.lanepe.2021.100056
Winkler, E. S., Gilchuk, P., Yu, J., Bailey, A. L., Chen, R. E., Chong, Z., et al. (2021). Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 184, 1804–1820.e16. e1816. doi:10.1016/j.cell.2021.02.026
Wölfel, R., Corman, V. M., Guggemos, W., Seilmaier, M., Zange, S., Müller, M. A., et al. (2020). Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469. doi:10.1038/s41586-020-2196-x
Wolter, N., Jassat, W., Walaza, S., Welch, R., Moultrie, H., Groome, M., et al. (2022). Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in south Africa: A data linkage study. Lancet 399, 437–446. doi:10.1016/s0140-6736(22)00017-4
Wong, J., Abdul Aziz, A. B. Z., Chaw, L., Mahamud, A., Griffith, M. M., Lo, Y. R., et al. (2020a). High proportion of asymptomatic and presymptomatic COVID-19 infections in air passengers to Brunei. J. Travel Med. 27, taaa066. doi:10.1093/jtm/taaa066
Wong, S. H., Lui, R. N., and Sung, J. J. (2020b). Covid-19 and the digestive system. J. Gastroenterology Hepatology 35, 744–748. doi:10.1111/jgh.15047
Woodruff, M. C., Ramonell, R. P., Nguyen, D. C., Cashman, K. S., Saini, A. S., Haddad, N. S., et al. (2020). Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 21, 1506–1516. doi:10.1038/s41590-020-00814-z
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263. doi:10.1126/science.abb2507
Wu, Y., Xu, X., Chen, Z., Duan, J., Hashimoto, K., Yang, L., et al. (2020). Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav. Immun. 87, 18–22. doi:10.1016/j.bbi.2020.03.031
Wu, Z., Hu, Y., Xu, M., Chen, Z., Yang, W., Jiang, Z., et al. (2021). Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine (CoronaVac) in healthy adults aged 60 years and older: A randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 21, 803–812. doi:10.1016/s1473-3099(20)30987-7
Wurm, J. P., Griese, M., Bahr, U., Held, M., Heckel, A., Karas, M., et al. (2012). Identification of the enzyme responsible for N1-methylation of pseudouridine 54 in archaeal tRNAs. Rna 18, 412–420. doi:10.1261/rna.028498.111
Xia, H., Cao, Z., Xie, X., Zhang, X., Chen, J. Y., Wang, H., et al. (2020). Evasion of type I interferon by SARS-CoV-2. Cell. Rep. 33, 108234. doi:10.1016/j.celrep.2020.108234
Xia, S., Zhang, Y., Wang, Y., Wang, H., Yang, Y., Gao, G. F., et al. (2021). Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: A randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 21, 39–51. doi:10.1016/s1473-3099(20)30831-8
Xia, S., Zhang, Y., Wang, Y., Wang, H., Yang, Y., Gao, G. F., et al. (2022). Safety and immunogenicity of an inactivated COVID-19 vaccine, BBIBP-CorV, in people younger than 18 years: A randomised, double-blind, controlled, phase 1/2 trial. Lancet Infect. Dis. 22, 196–208. doi:10.1016/s1473-3099(21)00462-x
Xia, X. (2021). Domains and functions of spike protein in sars-cov-2 in the context of vaccine design. Viruses 13, 109. doi:10.3390/v13010109
Xu, L., Zhang, H., and Wu, Y. (2014). Dendrimer advances for the central nervous system delivery of therapeutics. ACS Chem. Neurosci. 5, 2–13. doi:10.1021/cn400182z
Xu, J., Zhao, S., Teng, T., Abdalla, A. E., Zhu, W., Xie, L., et al. (2020a). Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses 12, 244. doi:10.3390/v12020244
Xu, R., Shi, M., Li, J., Song, P., and Li, N. (2020b). Corrigendum: Construction of SARS-CoV-2 virus-like particles by mammalian expression system. Front. Bioeng. Biotechnol. 8, 1026. doi:10.3389/fbioe.2020.01026
Xu, C., Lei, C., Hosseinpour, S., Ivanovski, S., Walsh, L. J., and Khademhosseini, A. (2022). Nanotechnology for the management of COVID-19 during the pandemic and in the post-pandemic era. Natl. Sci. Rev. 9, nwac124. doi:10.1093/nsr/nwac124
Yadav, P. D., Ella, R., Kumar, S., Patil, D. R., Mohandas, S., Shete, A. M., et al. (2021). Immunogenicity and protective efficacy of inactivated SARS-CoV-2 vaccine candidate, BBV152 in rhesus macaques. Nat. Commun. 12, 1386. doi:10.1038/s41467-021-21639-w
Yakovleva, A., Kovalenko, G., Redlinger, M., Liulchuk, M. G., Bortz, E., Zadorozhna, V. I., et al. (2021). Tracking SARS-COV-2 variants using nanopore sequencing in Ukraine in summer 2021. Res. Sq. 12, 15749. doi:10.21203/rs.3.rs-1044446/v1
Yakovleva, A., Kovalenko, G., Redlinger, M., Liulchuk, M. G., Bortz, E., Zadorozhna, V. I., et al. (2022). Tracking SARS-COV-2 variants using Nanopore sequencing in Ukraine in 2021. Sci. Rep. 12, 15749. doi:10.1038/s41598-022-19414-y
Yamayoshi, S., Sakai-Tagawa, Y., Koga, M., Akasaka, O., Nakachi, I., Koh, H., et al. (2020). Comparison of rapid antigen tests for COVID-19. Viruses 12, 1420. doi:10.3390/v12121420
Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., and Zhou, Q. (2020a). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448. doi:10.1126/science.abb2762
Yan, X., Han, X., Fan, Y., Fang, Z., Long, D., and Zhu, Y. (2020b). Duration of SARS-CoV-2 viral RNA in asymptomatic carriers. Crit. Care (Houten). 24, 245. doi:10.1186/s13054-020-02952-0
Yan, D., Zhang, X., Chen, C., Jiang, D., Liu, X., Zhou, Y., et al. (2021). Characteristics of viral shedding time in SARS-CoV-2 infections: A systematic review and meta-analysis. Front. Public Health 9, 652842. doi:10.3389/fpubh.2021.652842
Yang, J., Wang, W., Chen, Z., Lu, S., Yang, F., Bi, Z., et al. (2020a). A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 586, 572–577. doi:10.1038/s41586-020-2599-8
Yang, X., Yu, Y., Xu, J., Shu, H., Xia, J., Liu, H., et al. (2020b). Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 8, 475–481. doi:10.1016/s2213-2600(20)30079-5
Yang, M., Li, L., Huang, T., Li, S., Zhang, M., Yang, Y., et al. (2021a). SARS-CoV-2 detected on environmental fomites for both asymptomatic and symptomatic patients with COVID-19. Am. J. Respir. Crit. Care Med. 203, 374–378. doi:10.1164/rccm.202006-2136le
Yang, S., Li, Y., Dai, L., Wang, J., He, P., Li, C., et al. (2021b). Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: Two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect. Dis. 21, 1107–1119. doi:10.1016/s1473-3099(21)00127-4
Ye, Z.-W., Ong, C. P., Tang, K., Fan, Y., Luo, C., Zhou, R., et al. (2022). Intranasal administration of a single dose of a candidate live attenuated vaccine derived from an NSP16-deficient SARS-CoV-2 strain confers sterilizing immunity in animals. Cell. Mol. Immunol. 19, 588–601. doi:10.1038/s41423-022-00855-4
Yen, D. a.-W., Cappellini, B., Yang, H.-P., and Gupta, S. (2021). Coping with coping: International migrants’ experiences of the covid-19 lockdown in the UK. Brit. J. Manage. 32, 1219–1241. doi:10.1111/1467-8551.12512
Yilmaz, I. C., Ipekoglu, E. M., Bulbul, A., Turay, N., Yildirim, M., Evcili, I., et al. (2022). Development and preclinical evaluation of virus-like particle vaccine against COVID-19 infection. Allergy 77, 258–270. doi:10.1111/all.15091
Yu, K. K., Fischinger, S., Smith, M. T., Atyeo, C., Cizmeci, D., Wolf, C. R., et al. (2021). Comorbid illnesses are associated with altered adaptive immune responses to SARS-CoV-2. JCI Insight 6, 146242. doi:10.1172/jci.insight.146242
Zanet, V., Vidic, J., Auger, S., Vizzini, P., Lippe, G., Iacumin, L., et al. (2019). Activity evaluation of pure and doped zinc oxide nanoparticles against bacterial pathogens and Saccharomyces cerevisiae. J. Appl. Microbiol. 127, 1391–1402. doi:10.1111/jam.14407
Zaneti, R. N., Girardi, V., Spilki, F. R., Mena, K., Westphalen, A. P. C., Da Costa Colares, E. R., et al. (2021). Quantitative microbial risk assessment of SARS-CoV-2 for workers in wastewater treatment plants. Sci. Total Environ. 754, 142163. doi:10.1016/j.scitotenv.2020.142163
Zeltins, A. (2013). Construction and characterization of virus-like particles: A review. Mol. Biotechnol. 53, 92–107. doi:10.1007/s12033-012-9598-4
Zha, L., Chang, X., Zhao, H., Mohsen, M. O., Hong, L., Zhou, Y., et al. (2021). Development of a vaccine against SARS-CoV-2 based on the receptor-binding domain displayed on virus-like particles. Vaccines (Basel) 9, 395. doi:10.3390/vaccines9040395
Zhang, X. S., and Duchaine, C. (2020). SARS-CoV-2 and health care worker protection in low-risk settings: A review of modes of transmission and a novel airborne model involving inhalable particles. Clin. Microbiol. Rev. 34, 001844. doi:10.1128/cmr.00184-20
Zhang, C., Shi, L., and Wang, F. S. (2020a). Liver injury in COVID-19: Management and challenges. Lancet Gastroenterol. Hepatol. 5, 428–430. doi:10.1016/s2468-1253(20)30057-1
Zhang, L., Beatty, A., Lu, L., Abdalrahman, A., Makris, T. M., Wang, G., et al. (2020b). Microfluidic-assisted polymer-protein assembly to fabricate homogeneous functionalnanoparticles. Mater. Sci. Eng. C 111, 110768. doi:10.1016/j.msec.2020.110768
Zhang, L., Jackson, C. B., Mou, H., Ojha, A., Peng, H., Quinlan, B. D., et al. (2020c). SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 11, 6013. doi:10.1038/s41467-020-19808-4
Zhang, Q., Bastard, P., Liu, Z., Le Pen, J., Moncada-Velez, M., Chen, J., et al. (2020d). Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370, eabd4570. doi:10.1126/science.abd4570
Zhang, Q., Xiang, R., Huo, S., Zhou, Y., Jiang, S., Wang, Q., et al. (2021a). Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct. Target. Ther. 6, 233. doi:10.1038/s41392-021-00653-w
Zhang, Y., Zeng, G., Pan, H., Li, C., Hu, Y., Chu, K., et al. (2021b). Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18-59 years: A randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 21, 181–192. doi:10.1016/s1473-3099(20)30843-4
Zhang, Q., Bastard, P., Cobat, A., and Casanova, J. L. (2022a). Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature 603, 587–598. doi:10.1038/s41586-022-04447-0
Zhang, Y., Almazi, J. G., Ong, H. X., Johansen, M. D., Ledger, S., Traini, D., et al. (2022b). Nanoparticle delivery platforms for RNAi therapeutics targeting COVID-19 disease in the respiratory tract. Int. J. Mol. Sci. 23, 2408. doi:10.3390/ijms23052408
Zhao, B. S., and He, C. (2015). Pseudouridine in a new era of RNA modifications. Cell. Res. 25, 153–154. doi:10.1038/cr.2014.143
Zhao, L., Seth, A., Wibowo, N., Zhao, C. X., Mitter, N., Yu, C., et al. (2014). Nanoparticle vaccines. Vaccine 32, 327–337. doi:10.1016/j.vaccine.2013.11.069
Zhao, J., Yuan, Q., Wang, H., Liu, W., Liao, X., Su, Y., et al. (2020). Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin. Infect. Dis. 71, 2027–2034. doi:10.1093/cid/ciaa344
Zheng, Y. Y., Ma, Y. T., Zhang, J. Y., and Xie, X. (2020). COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 17, 259–260. doi:10.1038/s41569-020-0360-5
Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., et al. (2020a). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273. doi:10.1038/s41586-020-2012-7
Zhou, R., Li, F., Chen, F., Liu, H., Zheng, J., Lei, C., et al. (2020b). Viral dynamics in asymptomatic patients with COVID-19. Int. J. Infect. Dis. 96, 288–290. doi:10.1016/j.ijid.2020.05.030
Zhu, M., Wang, R., and Nie, G. (2014). Applications of nanomaterials as vaccine adjuvants. Hum. Vaccin. Immunother. 10, 2761–2774. doi:10.4161/hv.29589
Zhu, F. C., Guan, X. H., Li, Y. H., Huang, J. Y., Jiang, T., Hou, L. H., et al. (2020a). Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 396, 479–488. doi:10.1016/s0140-6736(20)31605-6
Zhu, F. C., Li, Y. H., Guan, X. H., Hou, L. H., Wang, W. J., Li, J. X., et al. (2020b). Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: A dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 395, 1845–1854. doi:10.1016/s0140-6736(20)31208-3
Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., et al. (2020c). A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. Overseas. Ed. 382, 727–733. doi:10.1056/nejmoa2001017
Zhu, X., Wang, X., Han, L., Chen, T., Wang, L., Li, H., et al. (2020d). Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. X. 166, 112437. doi:10.1016/j.bios.2020.112437
Zielińska, A., Carreiró, F., Oliveira, A. M., Neves, A., Pires, B., Venkatesh, D. N., et al. (2020). Polymeric nanoparticles: Production, characterization, toxicology and ecotoxicology. Molecules 25, 3731. doi:10.3390/molecules25163731
Znaidia, M., Demeret, C., Van Der Werf, S., and Komarova, A. V. (2022). Characterization of SARS-CoV-2 evasion: Interferon pathway and therapeutic options. Viruses 14, 1247. doi:10.3390/v14061247
Zou, L., Ruan, F., Huang, M., Liang, L., Huang, H., Hong, Z., et al. (2020). SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. Overseas. Ed. 382, 1177–1179. doi:10.1056/nejmc2001737
Keywords: nanoreagents, personal protective equipment, vaccines, lipid nanoparticles, modified mRNA, recombinant proteins, surveillance and diagnosis, genome sequencing
Citation: Valenzuela-Fernández A, Cabrera-Rodriguez R, Ciuffreda L, Perez-Yanes S, Estevez-Herrera J, González-Montelongo R, Alcoba-Florez J, Trujillo-González R, García-Martínez de Artola D, Gil-Campesino H, Díez-Gil O, Lorenzo-Salazar JM, Flores C and Garcia-Luis J (2022) Nanomaterials to combat SARS-CoV-2: Strategies to prevent, diagnose and treat COVID-19. Front. Bioeng. Biotechnol. 10:1052436. doi: 10.3389/fbioe.2022.1052436
Received: 23 September 2022; Accepted: 09 November 2022;
Published: 25 November 2022.
Edited by:
Donato Zipeto, University of Verona, ItalyReviewed by:
Massimiliano Perduca, University of Verona, ItalyGiuseppe Bardi, Italian Institute of Technology (IIT), Italy
Copyright © 2022 Valenzuela-Fernández, Cabrera-Rodriguez, Ciuffreda, Perez-Yanes, Estevez-Herrera, González-Montelongo, Alcoba-Florez, Trujillo-González, García-Martínez de Artola, Gil-Campesino, Díez-Gil, Lorenzo-Salazar, Flores and Garcia-Luis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Agustín Valenzuela-Fernández, avalenzu@ull.edu.es
 Agustín Valenzuela-Fernández
Agustín Valenzuela-Fernández Romina Cabrera-Rodriguez
Romina Cabrera-Rodriguez Laura Ciuffreda
Laura Ciuffreda Silvia Perez-Yanes
Silvia Perez-Yanes Judith Estevez-Herrera1
Judith Estevez-Herrera1  Rafaela González-Montelongo
Rafaela González-Montelongo Julia Alcoba-Florez
Julia Alcoba-Florez Carlos Flores
Carlos Flores